Q42521
Gene name |
GAD1 (GAD, GDH1) |
Protein name |
Glutamate decarboxylase 1 |
Names |
GAD 1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G17330 |
EC number |
4.1.1.15: Carboxy-lyases |
Protein Class |
|
Descriptions
(Annotation from UniProt)
The C-terminus region binds calmodulin in a calcium-dependent fashion and contains probably an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
469-502 (Calmodulin-binding domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
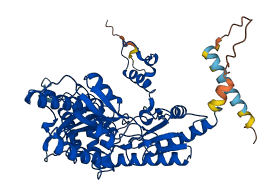
Autoinhibited structure

Activated structure

2 structures for Q42521
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3HBX | X-ray | 267 A | A/B/C/D/E/F | 1-502 | PDB |
| AF-Q42521-F1 | Predicted | AlphaFoldDB |
12 variants for Q42521
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH03067908 | 26 | S>P | No | 1000Genomes | |
| ENSVATH10806110 | 154 | K>Q | No | 1000Genomes | |
| tmp_5_5713195_G_A | 191 | D>N | No | 1000Genomes | |
| ENSVATH06981615 | 201 | E>D | No | 1000Genomes | |
| ENSVATH10806111 | 223 | L>V | No | 1000Genomes | |
| tmp_5_5713312_G_A | 230 | E>K | No | 1000Genomes | |
| tmp_5_5713321_A_C | 233 | K>Q | No | 1000Genomes | |
| ENSVATH10806113 | 310 | D>N | No | 1000Genomes | |
| ENSVATH00627149 | 363 | R>K | No | 1000Genomes | |
| ENSVATH00627150 | 370 | D>N | No | 1000Genomes | |
| ENSVATH00627151 | 413 | H>R | No | 1000Genomes | |
| tmp_5_5714787_C_T | 486 | T>I | No | 1000Genomes |
No associated diseases with Q42521
No regional properties for Q42521
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q42521 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | 4.1.1.15 | Carboxy-lyases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| glutamate decarboxylase activity | Catalysis of the reaction: L-glutamate = 4-aminobutanoate + CO2. |
| pyridoxal phosphate binding | Binding to pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| glutamate catabolic process | The chemical reactions and pathways resulting in the breakdown of glutamate, the anion of 2-aminopentanedioic acid. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q04792 | GAD1 | Glutamate decarboxylase | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P69908 | gadA | Glutamate decarboxylase alpha | Escherichia coli (strain K12) | PR |
| P69910 | gadB | Glutamate decarboxylase beta | Escherichia coli (strain K12) | EV |
| Q9LSH2 | GAD5 | Glutamate decarboxylase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZPS3 | GAD4 | Glutamate decarboxylase 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P54767 | Glutamate decarboxylase | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVLSHAVSES | DVSVHSTFAS | RYVRTSLPRF | KMPENSIPKE | AAYQIINDEL | MLDGNPRLNL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ASFVTTWMEP | ECDKLIMSSI | NKNYVDMDEY | PVTTELQNRC | VNMIAHLFNA | PLEEAETAVG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VGTVGSSEAI | MLAGLAFKRK | WQNKRKAEGK | PVDKPNIVTG | ANVQVCWEKF | ARYFEVELKE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VKLSEGYYVM | DPQQAVDMVD | ENTICVAAIL | GSTLNGEFED | VKLLNDLLVE | KNKETGWDTP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IHVDAASGGF | IAPFLYPELE | WDFRLPLVKS | INVSGHKYGL | VYAGIGWVIW | RNKEDLPEEL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IFHINYLGAD | QPTFTLNFSK | GSSQVIAQYY | QLIRLGHEGY | RNVMENCREN | MIVLREGLEK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TERFNIVSKD | EGVPLVAFSL | KDSSCHTEFE | ISDMLRRYGW | IVPAYTMPPN | AQHITVLRVV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| IREDFSRTLA | ERLVIDIEKV | MRELDELPSR | VIHKISLGQE | KSESNSDNLM | VTVKKSDIDK |
| 490 | 500 | ||||
| QRDIITGWKK | FVADRKKTSG | IC |