Q3ZC27
Gene name |
RAB19 |
Protein name |
Ras-related protein Rab-19 |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:506911 |
EC number |
|
Protein Class |
|
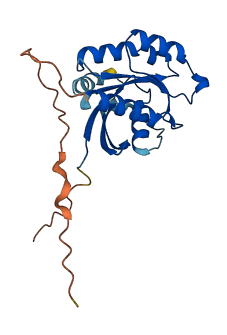
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q3ZC27
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q3ZC27-F1 | Predicted | AlphaFoldDB |
49 variants for Q3ZC27
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs525467856 | 9 | T>A | No | EVA | |
| rs468643709 | 32 | C>Y | No | EVA | |
| rs525601458 | 43 | M>T | No | EVA | |
| rs478420855 | 52 | V>A | No | EVA | |
| rs440764013 | 53 | D>A | No | EVA | |
| rs458316997 | 54 | F>I | No | EVA | |
| rs478294534 | 54 | F>Y | No | EVA | |
| rs716879743 | 57 | R>H | No | EVA | |
| rs461329441 | 74 | A>V | No | EVA | |
| rs481430161 | 77 | E>Q | No | EVA | |
| rs456936809 | 81 | T>S | No | EVA | |
| rs449109978 | 83 | T>P | No | EVA | |
| rs434596171 | 97 | Y>S | No | EVA | |
| rs444991674 | 98 | D>G | No | EVA | |
| rs464929188 | 100 | T>P | No | EVA | |
| rs464929188 | 100 | T>S | No | EVA | |
| rs436853511 | 104 | T>S | No | EVA | |
| rs470747747 | 136 | E>V | No | EVA | |
| rs462580938 | 152 | Y>C | No | EVA | |
| rs482673906 | 158 | L>R | No | EVA | |
| rs479097523 | 167 | N>H | No | EVA | |
| rs447821612 | 168 | I>S | No | EVA | |
| rs464610222 | 176 | A>P | No | EVA | |
| rs470270172 | 179 | L>R | No | EVA | |
| rs450169387 | 179 | L>V | No | EVA | |
| rs435619447 | 180 | M>R | No | EVA | |
| rs455715642 | 181 | A>P | No | EVA | |
| rs479317256 | 183 | H>P | No | EVA | |
| rs433888893 | 190 | E>* | No | EVA | |
| rs524921982 | 193 | P>T | No | EVA | |
| rs453985808 | 195 | S>N | No | EVA | |
| rs453985808 | 195 | S>T | No | EVA | |
| rs470814764 | 196 | L>R | No | EVA | |
| rs456259896 | 198 | L>R | No | EVA | |
| rs476295486 | 199 | E>A | No | EVA | |
| rs476295486 | 199 | E>G | No | EVA | |
| rs441600854 | 200 | S>A | No | EVA | |
| rs441600854 | 200 | S>T | No | EVA | |
| rs461555224 | 205 | M>I | No | EVA | |
| rs1116811577 | 207 | P>L | No | EVA | |
| rs471976362 | 209 | P>A | No | EVA | |
| rs458329506 | 210 | R>T | No | EVA | |
| rs450192317 | 211 | E>D | No | EVA | |
| rs478501619 | 211 | E>Q | No | EVA | |
| rs463900991 | 212 | K>Q | No | EVA | |
| rs449374508 | 214 | Q>H | No | EVA | |
| rs480754954 | 214 | Q>P | No | EVA | |
| rs466183771 | 215 | C>G | No | EVA | |
| rs434728874 | 216 | T>A | No | EVA |
No associated diseases with Q3ZC27
1 regional properties for Q3ZC27
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 16 - 175 | IPR005225 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome assembly | The formation of a double membrane-bounded structure, the autophagosome, that occurs when a specialized membrane sac, called the isolation membrane, starts to enclose a portion of the cytoplasm. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
7 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5ZHV1 | RAB33B | Ras-related protein Rab-33B | Gallus gallus (Chicken) | PR |
| Q9H082 | RAB33B | Ras-related protein Rab-33B | Homo sapiens (Human) | PR |
| A4D1S5 | RAB19 | Ras-related protein Rab-19 | Homo sapiens (Human) | PR |
| O35963 | Rab33b | Ras-related protein Rab-33B | Mus musculus (Mouse) | PR |
| P35294 | Rab19 | Ras-related protein Rab-19 | Mus musculus (Mouse) | PR |
| Q5M7U5 | Rab19 | Ras-related protein Rab-19 | Rattus norvegicus (Rat) | PR |
| Q20365 | rab-33 | Ras-related protein Rab-33 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQFSSSARTV | DENFDYLFKI | ILIGDSNVGK | TCVVQHFKSG | VYMEAQQNTI | GVDFTVRALE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IDGKKVKMQV | WDTAGQERFR | TITQSYYRSA | HAAIIAYDLT | RRSTFESVPH | WIHEIEKYGA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ANLVIMLIGN | KCDLWEKRHV | LFEDACILAE | KYGLLAVLET | SAKESKNIDE | VFVLMARELM |
| 190 | 200 | 210 | |||
| ARHSLPLYGE | GAPGSLPLES | TPVLMAPAPR | EKNQCTC |