Q3SYZ2
Gene name |
MAPKAPK3 |
Protein name |
MAP kinase-activated protein kinase 3 |
Names |
MAPK-activated protein kinase 3 , MAPKAP kinase 3 , MAPKAP-K3 , MAPKAPK-3 , MK-3 , EC 2.7.11.1 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:615215 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
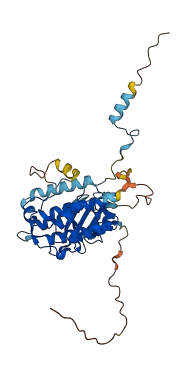
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
5-306 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
188-209 (Activation loop from InterPro)
Target domain |
45-306 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
188-209 (Activation loop from InterPro)
Target domain |
45-306 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Seternes OM et al. (2002) "Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress", Molecular and cellular biology, 22, 6931-45
- Engel K et al. (1998) "Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation", The EMBO journal, 17, 3363-71
Autoinhibited structure

Activated structure

1 structures for Q3SYZ2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q3SYZ2-F1 | Predicted | AlphaFoldDB |
108 variants for Q3SYZ2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs449498285 | 2 | D>A | No | EVA | |
| rs480692966 | 3 | V>G | No | EVA | |
| rs463704334 | 4 | E>D | No | EVA | |
| rs443748584 | 9 | Q>P | No | EVA | |
| rs438574813 | 11 | G>S | No | EVA | |
| rs473295106 | 12 | P>T | No | EVA | |
| rs442681455 | 16 | S>A | No | EVA | |
| rs436881348 | 18 | V>G | No | EVA | |
| rs456855620 | 18 | V>I | No | EVA | |
| rs451795984 | 19 | P>T | No | EVA | |
| rs434999577 | 20 | C>W | No | EVA | |
| rs473449610 | 21 | G>R | No | EVA | |
| rs449600392 | 28 | P>R | No | EVA | |
| rs480957243 | 29 | A>G | No | EVA | |
| rs449972995 | 34 | R>W | No | EVA | |
| rs458181950 | 36 | P>T | No | EVA | |
| rs479646892 | 37 | K>N | No | EVA | |
| rs459461361 | 39 | Y>* | No | EVA | |
| rs455065201 | 40 | A>G | No | EVA | |
| rs474079917 | 43 | D>E | No | EVA | |
| rs443141859 | 44 | D>E | No | EVA | |
| rs456936280 | 44 | D>G | No | EVA | |
| rs471207324 | 46 | Q>K | No | EVA | |
| rs451338472 | 47 | L>S | No | EVA | |
| rs434434207 | 51 | V>G | No | EVA | |
| rs466414090 | 60 | V>G | No | EVA | |
| rs455831767 | 61 | L>R | No | EVA | |
| rs435904999 | 68 | T>S | No | EVA | |
| rs450068590 | 70 | Q>H | No | EVA | |
| rs433199930 | 71 | K>T | No | EVA | |
| rs464547462 | 73 | A>S | No | EVA | |
| rs447819872 | 75 | K>T | No | EVA | |
| rs134288208 | 86 | E>G | No | EVA | |
| rs132904374 | 87 | V>A | No | EVA | |
| rs132904374 | 87 | V>G | No | EVA | |
| rs460473582 | 87 | V>L | No | EVA | |
| rs135090864 | 88 | D>A | No | EVA | |
| rs474621418 | 90 | H>P | No | EVA | |
| rs454489934 | 93 | A>P | No | EVA | |
| rs475552971 | 96 | G>C | No | EVA | |
| rs432612053 | 99 | I>L | No | EVA | |
| rs447121417 | 100 | V>G | No | EVA | |
| rs466981343 | 100 | V>L | No | EVA | |
| rs436619672 | 101 | R>C | No | EVA | |
| rs467611360 | 102 | I>L | No | EVA | |
| rs450752771 | 103 | L>P | No | EVA | |
| rs459024788 | 104 | D>E | No | EVA | |
| rs482269105 | 104 | D>G | No | EVA | |
| rs445315267 | 105 | V>D | No | EVA | |
| rs445315267 | 105 | V>G | No | EVA | |
| rs477227114 | 106 | Y>F | No | EVA | |
| rs460307955 | 107 | E>G | No | EVA | |
| rs440349668 | 111 | H>P | No | EVA | |
| rs470558967 | 165 | A>D | No | EVA | |
| rs453849517 | 170 | K>N | No | EVA | |
| rs443054245 | 172 | E>A | No | EVA | |
| rs474393488 | 209 | Y>D | No | EVA | |
| rs451747169 | 219 | E>D | No | EVA | |
| rs433222383 | 231 | G>C | No | EVA | |
| rs455074304 | 239 | C>R | No | EVA | |
| rs438157829 | 241 | F>S | No | EVA | |
| rs452677509 | 247 | N>D | No | EVA | |
| rs432369996 | 247 | N>K | No | EVA | |
| rs466953259 | 249 | G>S | No | EVA | |
| rs446911549 | 252 | I>L | No | EVA | |
| rs467839274 | 259 | R>S | No | EVA | |
| rs448329988 | 262 | L>Q | No | EVA | |
| rs482800164 | 267 | F>V | No | EVA | |
| rs439499396 | 272 | W>G | No | EVA | |
| rs477060648 | 275 | V>G | No | EVA | |
| rs460113799 | 277 | E>G | No | EVA | |
| rs876281510 | 311 | V>G | No | EVA | |
| rs465870187 | 311 | V>L | No | EVA | |
| rs876177939 | 312 | V>G | No | EVA | |
| rs875993360 | 315 | T>P | No | EVA | |
| rs449443523 | 318 | H>P | No | EVA | |
| rs876217918 | 320 | A>P | No | EVA | |
| rs475010084 | 335 | E>G | No | EVA | |
| rs461336417 | 338 | T>P | No | EVA | |
| rs444587658 | 339 | S>R | No | EVA | |
| rs475944648 | 341 | L>V | No | EVA | |
| rs452710715 | 341 | L>W | No | EVA | |
| rs432375181 | 343 | T>N | No | EVA | |
| rs453384173 | 344 | M>I | No | EVA | |
| rs473360348 | 344 | M>L | No | EVA | |
| rs473360348 | 344 | M>V | No | EVA | |
| rs436439020 | 346 | V>A | No | EVA | |
| rs467862275 | 347 | D>A | No | EVA | |
| rs454770295 | 348 | Y>* | No | EVA | |
| rs445858535 | 351 | V>A | No | EVA | |
| rs445858535 | 351 | V>G | No | EVA | |
| rs469336231 | 351 | V>L | No | EVA | |
| rs466476253 | 352 | K>T | No | EVA | |
| rs446507411 | 353 | I>S | No | EVA | |
| rs460874504 | 355 | D>G | No | EVA | |
| rs480961661 | 355 | D>H | No | EVA | |
| rs480961661 | 355 | D>N | No | EVA | |
| rs444634999 | 356 | L>P | No | EVA | |
| rs482447558 | 357 | K>E | No | EVA | |
| rs459272541 | 358 | T>A | No | EVA | |
| rs459272541 | 358 | T>S | No | EVA | |
| rs439038508 | 360 | N>D | No | EVA | |
| rs439038508 | 360 | N>Y | No | EVA | |
| rs473438913 | 361 | N>S | No | EVA | |
| rs442844009 | 363 | L>P | No | EVA | |
| rs437814241 | 379 | Q>K | No | EVA | |
| rs469124066 | 380 | G>A | No | EVA | |
| rs452380808 | 382 | N>H | No | EVA |
No associated diseases with Q3SYZ2
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein serine/threonine kinase activity | Calcium-dependent catalysis of the reactions |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions |
| mitogen-activated protein kinase binding | Binding to a mitogen-activated protein kinase. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| macropinocytosis | An endocytosis process that results in the uptake of liquid material by cells from their external environment by the 'ruffling' of the cell membrane to form heterogeneously sized intracellular vesicles called macropinosomes, which can be up to 5 micrometers in size. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| response to cytokine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor of a target cell. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P49071 | MAPk-Ak2 | MAP kinase-activated protein kinase 2 | Drosophila melanogaster (Fruit fly) | SS |
| P49137 | MAPKAPK2 | MAP kinase-activated protein kinase 2 | Homo sapiens (Human) | EV |
| Q8IW41 | MAPKAPK5 | MAP kinase-activated protein kinase 5 | Homo sapiens (Human) | SS |
| Q16644 | MAPKAPK3 | MAP kinase-activated protein kinase 3 | Homo sapiens (Human) | SS |
| O54992 | Mapkapk5 | MAP kinase-activated protein kinase 5 | Mus musculus (Mouse) | EV |
| P49138 | Mapkapk2 | MAP kinase-activated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q3UMW7 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Mus musculus (Mouse) | SS |
| Q66H84 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Rattus norvegicus (Rat) | SS |
| Q965G5 | mak-2 | MAP kinase-activated protein kinase mak-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDVETAEEQG | GPAPPSGVPC | GPCSAGAPAL | GGRREPKKYA | VTDDYQLSKQ | VLGLGVNGKV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LECFHRRTGQ | KCALKLLYDS | PKARQEVDHH | WQASGGPHIV | RILDVYENMH | HSKRCLLIIM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ECMEGGELFS | RIQERGDQAF | TEREAAEIMR | DIGTAIQFLH | SRNIAHRDVK | PENLLYTSKD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KDAVLKLTDF | GFAKETTQNA | LQTPCYTPYY | VAPEVLGPEK | YDKSCDMWSL | GVIMYILLCG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FPPFYSNTGQ | AISPGMKRRI | RLGQYGFPSP | EWSEVSEDAK | QLIRLLLKTD | PTERLTITQF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| MNHPWINQSM | VVPQTPLHTA | RVLQEDRDHW | DEVKEEMTSA | LATMRVDYDQ | VKIKDLKTSN |
| 370 | 380 | ||||
| NRLLNKRRKK | QAGSSSGSQG | CNNQ |