Q3MHP2
Gene name |
RAB11B |
Protein name |
Ras-related protein Rab-11B |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:532723 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
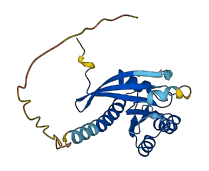
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q3MHP2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q3MHP2-F1 | Predicted | AlphaFoldDB |
116 variants for Q3MHP2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs434225351 | 6 | D>N | No | EVA | |
| rs454424445 | 12 | F>S | No | EVA | |
| rs453065720 | 14 | V>E | No | EVA | |
| rs438529334 | 15 | V>A | No | EVA | |
| rs438529334 | 15 | V>G | No | EVA | |
| rs473122270 | 15 | V>L | No | EVA | |
| rs458501755 | 17 | I>M | No | EVA | |
| rs475414047 | 18 | G>V | No | EVA | |
| rs443860010 | 19 | D>A | No | EVA | |
| rs480759240 | 21 | G>A | No | EVA | |
| rs459945985 | 22 | V>E | No | EVA | |
| rs459945985 | 22 | V>G | No | EVA | |
| rs445302405 | 23 | G>A | No | EVA | |
| rs465338730 | 24 | K>* | No | EVA | |
| rs465338730 | 24 | K>E | No | EVA | |
| rs437216651 | 24 | K>T | No | EVA | |
| rs450947387 | 26 | N>D | No | EVA | |
| rs467653823 | 26 | N>I | No | EVA | |
| rs436316467 | 26 | N>K | No | EVA | |
| rs467653823 | 26 | N>S | No | EVA | |
| rs453078445 | 32 | T>P | No | EVA | |
| rs473186363 | 33 | R>P | No | EVA | |
| rs432014688 | 35 | E>D | No | EVA | |
| rs452125235 | 41 | K>N | No | EVA | |
| rs443984841 | 46 | V>A | No | EVA | |
| rs460823448 | 47 | E>D | No | EVA | |
| rs474308445 | 48 | F>L | No | EVA | |
| rs443102393 | 50 | T>P | No | EVA | |
| rs480088263 | 57 | G>C | No | EVA | |
| rs482350103 | 66 | D>A | No | EVA | |
| rs458961226 | 66 | D>N | No | EVA | |
| rs450910386 | 69 | G>S | No | EVA | |
| rs467815536 | 70 | Q>K | No | EVA | |
| rs446713507 | 72 | R>P | No | EVA | |
| rs481265733 | 72 | R>S | No | EVA | |
| rs466749354 | 74 | R>P | No | EVA | |
| rs452305583 | 77 | T>A | No | EVA | |
| rs468898851 | 78 | S>P | No | EVA | |
| rs433205995 | 80 | Y>D | No | EVA | |
| rs456488507 | 80 | Y>S | No | EVA | |
| rs469981038 | 83 | G>A | No | EVA | |
| rs435553878 | 85 | V>L | No | EVA | |
| rs455686535 | 86 | G>S | No | EVA | |
| rs472517640 | 90 | V>L | No | EVA | |
| rs434724361 | 91 | Y>F | No | EVA | |
| rs451433809 | 124 | N>K | No | EVA | |
| rs471611272 | 126 | S>G | No | EVA | |
| rs443318415 | 127 | D>A | No | EVA | |
| rs463429601 | 130 | H>P | No | EVA | |
| rs459999679 | 152 | E>G | No | EVA | |
| rs479863443 | 154 | S>P | No | EVA | |
| rs449730644 | 171 | E>A | No | EVA | |
| rs463245993 | 171 | E>D | No | EVA | |
| rs449730644 | 171 | E>V | No | EVA | |
| rs483351645 | 172 | I>F | No | EVA | |
| rs483351645 | 172 | I>L | No | EVA | |
| rs468760135 | 172 | I>M | No | EVA | |
| rs448621029 | 172 | I>S | No | EVA | |
| rs448621029 | 172 | I>T | No | EVA | |
| rs483351645 | 172 | I>V | No | EVA | |
| rs434017064 | 173 | Y>H | No | EVA | |
| rs434017064 | 173 | Y>N | No | EVA | |
| rs447721275 | 173 | Y>S | No | EVA | |
| rs433133092 | 174 | R>C | No | EVA | |
| rs456416911 | 174 | R>H | No | EVA | |
| rs456416911 | 174 | R>L | No | EVA | |
| rs456416911 | 174 | R>P | No | EVA | |
| rs433133092 | 174 | R>S | No | EVA | |
| rs476539121 | 175 | I>F | No | EVA | |
| rs476539121 | 175 | I>L | No | EVA | |
| rs435480860 | 175 | I>T | No | EVA | |
| rs476539121 | 175 | I>V | No | EVA | |
| rs440914665 | 176 | V>A | No | EVA | |
| rs440914665 | 176 | V>E | No | EVA | |
| rs472396479 | 176 | V>L | No | EVA | |
| rs472396479 | 176 | V>M | No | EVA | |
| rs471525616 | 177 | S>A | No | EVA | |
| rs443242750 | 177 | S>L | No | EVA | |
| rs471525616 | 177 | S>P | No | EVA | |
| rs462326594 | 178 | Q>H | No | EVA | |
| rs483285883 | 178 | Q>K | No | EVA | |
| rs448697490 | 178 | Q>P | No | EVA | |
| rs478993533 | 179 | K>Q | No | EVA | |
| rs447831366 | 179 | K>T | No | EVA | |
| rs464485644 | 180 | Q>K | No | EVA | |
| rs433294957 | 180 | Q>L | No | EVA | |
| rs433294957 | 180 | Q>R | No | EVA | |
| rs470176351 | 181 | I>L | No | EVA | |
| rs455574162 | 181 | I>M | No | EVA | |
| rs435437488 | 181 | I>N | No | EVA | |
| rs435437488 | 181 | I>S | No | EVA | |
| rs435437488 | 181 | I>T | No | EVA | |
| rs472515715 | 182 | A>P | No | EVA | |
| rs472515715 | 182 | A>T | No | EVA | |
| rs471445980 | 183 | D>G | No | EVA | |
| rs451557965 | 183 | D>H | No | EVA | |
| rs463331656 | 184 | R>P | No | EVA | |
| rs473824228 | 185 | A>E | No | EVA | |
| rs442437714 | 186 | A>E | No | EVA | |
| rs442437714 | 186 | A>G | No | EVA | |
| rs479155108 | 187 | H>Q | No | EVA | |
| rs441366035 | 188 | D>A | No | EVA | |
| rs478065562 | 189 | E>G | No | EVA | |
| rs458179445 | 189 | E>Q | No | EVA | |
| rs450114381 | 190 | S>T | No | EVA | |
| rs480652456 | 191 | P>L | No | EVA | |
| rs470169448 | 191 | P>T | No | EVA | |
| rs449301333 | 198 | I>T | No | EVA | |
| rs466054683 | 199 | S>N | No | EVA | |
| rs451480013 | 202 | P>T | No | EVA | |
| rs465179114 | 204 | T>P | No | EVA | |
| rs436860073 | 205 | D>A | No | EVA | |
| rs456958104 | 210 | N>I | No | EVA | |
| rs473962703 | 212 | L>Q | No | EVA | |
| rs133469651 | 214 | C>S | No | EVA | |
| rs442384947 | 216 | Q>K | No | EVA |
No associated diseases with Q3MHP2
1 regional properties for Q3MHP2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 10 - 168 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| anchoring junction | A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| phagocytic vesicle | A membrane-bounded intracellular vesicle that arises from the ingestion of particulate material by phagocytosis. |
| phagocytic vesicle membrane | The lipid bilayer surrounding a phagocytic vesicle. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
| recycling endosome membrane | The lipid bilayer surrounding a recycling endosome. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
| synaptic vesicle membrane | The lipid bilayer surrounding a synaptic vesicle. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein activity | A molecular function regulator that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular processes. Intrinsic GTPase activity returns the G protein to its GDP-bound state. The return to the GDP-bound state can be accelerated by the action of a GTPase-activating protein (GAP). |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to acidic pH | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a pH stimulus with pH < 7. pH is a measure of the acidity or basicity of an aqueous solution. |
| constitutive secretory pathway | A process of exocytosis found in all eukaryotic cells, in which transport vesicles destined for the plasma membrane leave the trans-Golgi network in a steady stream. Upon exocytosis, the membrane proteins and lipids in these vesicles provide new components for the plasma membrane, and the soluble proteins inside the vesicles are released into the extracellular space. |
| endocytic recycling | The directed movement of membrane-bounded vesicles from endosomes back to the plasma membrane, a trafficking pathway that promotes the recycling of internalized transmembrane proteins. |
| exocytosis | A process of secretion by a cell that results in the release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle. Exocytosis can occur either by full fusion, when the vesicle collapses into the plasma membrane, or by a kiss-and-run mechanism that involves the formation of a transient contact, a pore, between a granule (for exemple of chromaffin cells) and the plasma membrane. The latter process most of the time leads to only partial secretion of the granule content. Exocytosis begins with steps that prepare vesicles for fusion with the membrane (tethering and docking) and ends when molecules are secreted from the cell. |
| insulin secretion involved in cellular response to glucose stimulus | The regulated release of proinsulin from secretory granules (B granules) in the B cells of the pancreas; accompanied by cleavage of proinsulin to form mature insulin, in response to a glucose stimulus. |
| melanosome transport | The directed movement of melanosomes into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| receptor recycling | The process that results in the return of receptor molecules to an active state and an active cellular location after they have been stimulated by a ligand. An active state is when the receptor is ready to receive a signal. |
| regulated exocytosis | A process of exocytosis in which soluble proteins and other substances are initially stored in secretory vesicles for later release. It is found mainly in cells that are specialized for secreting products such as hormones, neurotransmitters, or digestive enzymes rapidly on demand. |
| regulation of anion transport | Any process that modulates the frequency, rate or extent of the directed movement of anions, atoms or small molecules with a net negative charge into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of endocytic recycling | Any process that modulates the frequency, rate or extent of endocytic recycling. |
| regulation of protein localization to cell surface | Any process that modulates the frequency, rate or extent of protein localization to the cell surface. |
| transferrin transport | The directed movement of transferrin into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
16 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2TBH7 | RAB4A | Ras-related protein Rab-4A | Bos taurus (Bovine) | PR |
| Q2TA29 | RAB11A | Ras-related protein Rab-11A | Bos taurus (Bovine) | PR |
| Q5ZJN2 | RAB11A | Ras-related protein Rab-11A | Gallus gallus (Chicken) | PR |
| P62491 | RAB11A | Ras-related protein Rab-11A | Homo sapiens (Human) | PR |
| Q15907 | RAB11B | Ras-related protein Rab-11B | Homo sapiens (Human) | PR |
| P62492 | Rab11a | Ras-related protein Rab-11A | Mus musculus (Mouse) | PR |
| P46638 | Rab11b | Ras-related protein Rab-11B | Mus musculus (Mouse) | PR |
| Q52NJ1 | RAB11A | Ras-related protein Rab-11A | Sus scrofa (Pig) | PR |
| P62494 | Rab11a | Ras-related protein Rab-11A | Rattus norvegicus (Rat) | PR |
| O35509 | Rab11b | Ras-related protein Rab-11B | Rattus norvegicus (Rat) | PR |
| Q9LNK1 | RABA3 | Ras-related protein RABA3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH50 | RABA4D | Ras-related protein RABA4d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FE79 | RABA4C | Ras-related protein RABA4c | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O04486 | RABA2A | Ras-related protein RABA2a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O49513 | RABA1E | Ras-related protein RABA1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LNW1 | RABA2B | Ras-related protein RABA2b | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGTRDDEYDY | LFKVVLIGDS | GVGKSNLLSR | FTRNEFNLES | KSTIGVEFAT | RSIQVDGKTI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KAQIWDTAGQ | ERYRAITSAY | YRGAVGALLV | YDIAKHLTYE | NVERWLKELR | DHADSNIVIM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LVGNKSDLRH | LRAVPTDEAR | AFAEKNNLSF | IETSALDSTN | VEEAFKNILT | EIYRIVSQKQ |
| 190 | 200 | 210 | |||
| IADRAAHDES | PGNNVVDISV | PPTTDGQKPN | KLQCCQNL |