Q3KK31
Gene name |
lapD |
Protein name |
Diguanylate cyclase/phosphodiesterase |
Names |
|
Species |
Pseudomonas fluorescens (strain Pf0-1) |
KEGG Pathway |
pfo:Pfl01_0131 |
EC number |
|
Protein Class |
CYCLIC DI-GMP PHOSPHODIESTERASE PDEF (PTHR33121) |
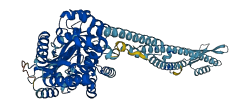
Descriptions
Bis-(39-59) cyclic dimeric guanosine monophosphate (c-d-GMP) is a monocyclic RNA dinucleotide controlling biofilm formation via a transmembrane receptor LapD, a periplasmic cysteine protease LapG, and surface adhesin protein LapA depending on the level of phosphate in the medium. When the phosphate level is low, LapD is held in the off state by interactions of their EAL domain (LapD 399-648), with the S helix (LapD 171-238) in HAMP domain and the GGDEF domain (LapD 238-399), which allows the proteolytic reaction of LapA on the cell surface, and consequently biofilm dispersal. Interaction of c-d-GMP to the phosphodiesterase binding site on the EAL domain is required to stimulate the receptor (LapD) by release the S helix and the GGDEF domain facilitating the formation of a trans-subunit dimer interface, the binding of LapG to LapD’s output domain, and consequently permitting the activation of LapA to form biofilm.
Autoinhibitory domains (AIDs)
Target domain |
399-648 (EAL domain) |
Relief mechanism |
Ligand binding |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure
Activated structure
No variants for Q3KK31
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q3KK31 | |||||
No associated diseases with Q3KK31
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR33121 | CYCLIC DI-GMP PHOSPHODIESTERASE PDEF |
| PANTHER Subfamily | PTHR33121:SF23 | CYCLIC DI-GMP PHOSPHODIESTERASE PDEB |
| PANTHER Protein Class | phosphodiesterase | |
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| nucleotide binding | Binding to a nucleotide, any compound consisting of a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the ribose or deoxyribose. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLFKQLLIA | ICLFLVVAFT | GSFMVSLESS | RTQYVNQLRS | HAQDAATALA | LSLTPNIDDP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AMVELLVSSI | FDSGYYSSIR | VVDLKTDQTI | VERNGIPAVT | NVPDWFVKLI | GLEPAGGDAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VSRGWEQAAR | VEVVSHPMFA | LAKLWQSALG | SLGWLLVCGA | VSAVLGALLL | RRQLKPLDYM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VKQSHAIARR | EFLSLPDLPR | TPELRRVVLA | MNQMVEKLKA | LFQEQAERSE | KLRTESYQDN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTGLANRRYF | EMQLNARVSN | PEQASSGYLL | LLRVKDLAGL | NQRLGGQRTD | ELLKAVGEQL |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SRECAKYPET | QNLVTRIRGG | EFAVLAPGMT | REEALQLAQS | LDSALSSLYA | TGATDVAAVA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SIGLAPFAHG | DSPQAVLSLG | DQALAQAEGQ | GEQNWACLDQ | SLVADVGDDH | HAWHRLLDQA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LNQRRFELFF | QPVVAAQDTQ | LVLHYKVLSR | LLDEQGQTIP | AGRFLPWLER | FGWTARLDRL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| MLERVLEQMA | GHEESLALNL | SSATLADPQA | LNKVFEILRA | HSNLGARLTL | EIGEEQLPEQ |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AVLEQLTRRL | RELGFSLSLQ | RFGGRFSMIG | NLARLGLAYL | KIDGSYIRAI | DQESDKRLFI |
| 610 | 620 | 630 | 640 | ||
| EAIQRAAHSI | DLPLIAERVE | TEGELSVIRE | MGLYGVQGQL | FGEPKPWG |