Q39032
Gene name |
PLC1 (ATHATPLC1G, At5g58670, MZN1.12) |
Protein name |
Phosphoinositide phospholipase C 1 |
Names |
Phosphoinositide phospholipase PLC1, AtPLC1, AtPLC1S, PI-PLC1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G58670 |
EC number |
3.1.4.11: Phosphoric diester hydrolases |
Protein Class |
|
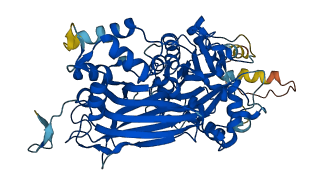
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q39032
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q39032-F1 | Predicted | AlphaFoldDB |
38 variants for Q39032
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_5_23706657_T_C | 32 | D>G | No | 1000Genomes | |
| ENSVATH12845239 | 38 | R>M | No | 1000Genomes | |
| ENSVATH12845238 | 43 | E>D | No | 1000Genomes | |
| tmp_5_23706590_T_A | 54 | E>D | No | 1000Genomes | |
| ENSVATH12845237 | 80 | I>M | No | 1000Genomes | |
| tmp_5_23706502_C_T | 84 | G>R | No | 1000Genomes | |
| ENSVATH00741260 | 87 | R>G | No | 1000Genomes | |
| ENSVATH07446266 | 97 | L>M | No | 1000Genomes | |
| ENSVATH07446265 | 119 | G>R | No | 1000Genomes | |
| tmp_5_23706277_T_C | 135 | I>V | No | 1000Genomes | |
| ENSVATH12845235 | 151 | L>I | No | 1000Genomes | |
| ENSVATH07446264 | 159 | G>E | No | 1000Genomes | |
| ENSVATH07446263 | 160 | K>R | No | 1000Genomes | |
| ENSVATH07446259 | 210 | K>R | No | 1000Genomes | |
| ENSVATH14639664 | 215 | V>M | No | 1000Genomes | |
| tmp_5_23705832_T_C | 229 | E>G | No | 1000Genomes | |
| tmp_5_23705748_G_A | 257 | T>I | No | 1000Genomes | |
| ENSVATH07446256 | 269 | T>I | No | 1000Genomes | |
| ENSVATH07446257 | 269 | T>S | No | 1000Genomes | |
| tmp_5_23705709_C_A | 270 | R>I | No | 1000Genomes | |
| ENSVATH07446255 | 288 | S>G | No | 1000Genomes | |
| tmp_5_23705567_C_T | 294 | R>K | No | 1000Genomes | |
| tmp_5_23705522_T_C | 309 | K>R | No | 1000Genomes | |
| tmp_5_23705507_C_T | 314 | G>E | No | 1000Genomes | |
| ENSVATH00741259 | 333 | A>T | No | 1000Genomes | |
| ENSVATH07446254 | 342 | K>R | No | 1000Genomes | |
| tmp_5_23705308_C_T | 380 | M>I | No | 1000Genomes | |
| tmp_5_23705140_A_T | 409 | L>M | No | 1000Genomes | |
| tmp_5_23705130_T_G | 412 | N>T | No | 1000Genomes | |
| tmp_5_23705110_A_C | 419 | F>V | No | 1000Genomes | |
| ENSVATH14639661 | 422 | C>F | No | 1000Genomes | |
| tmp_5_23704958_C_G | 441 | G>A | No | 1000Genomes | |
| tmp_5_23704948_C_G | 444 | M>I | No | 1000Genomes | |
| ENSVATH07446251 | 454 | Y>F | No | 1000Genomes | |
| ENSVATH14639659 | 464 | G>E | No | 1000Genomes | |
| tmp_5_23704728_G_A | 472 | T>I | No | 1000Genomes | |
| tmp_5_23704629_C_T | 505 | C>Y | No | 1000Genomes | |
| tmp_5_23704595_C_A | 516 | Q>H | No | 1000Genomes |
No associated diseases with Q39032
8 regional properties for Q39032
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 59 - 317 | IPR000719 |
| domain | EF-hand domain | 360 - 431 | IPR002048-1 |
| domain | EF-hand domain | 432 - 504 | IPR002048-2 |
| active_site | Serine/threonine-protein kinase, active site | 179 - 191 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 65 - 88 | IPR017441 |
| binding_site | EF-Hand 1, calcium-binding site | 409 - 421 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 445 - 457 | IPR018247-2 |
| binding_site | EF-Hand 1, calcium-binding site | 482 - 494 | IPR018247-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.11 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| metal ion binding | Binding to a metal ion. |
| phosphatidylinositol phospholipase C activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H(2)O = 1,2-diacylglycerol + 1D-myo-inositol 1,4,5-trisphosphate + H(+). |
| phospholipase C activity | Catalysis of the reaction: a phospholipid + H2O = 1,2-diacylglycerol + a phosphatidate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| abscisic acid-activated signaling pathway | The series of molecular signals generated by the binding of the plant hormone abscisic acid (ABA) to a receptor, and ending with modulation of a cellular process, e.g. transcription. |
| lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| response to abscisic acid | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an abscisic acid stimulus. |
| response to cold | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cold stimulus, a temperature stimulus below the optimal temperature for that organism. |
| response to salt stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating an increase or decrease in the concentration of salt (particularly but not exclusively sodium and chloride ions) in the environment. |
| response to water deprivation | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a water deprivation stimulus, prolonged deprivation of water. |
43 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32383 | PLC1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P08487 | PLCG1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Bos taurus (Bovine) | EV SS |
| P10895 | PLCD1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Bos taurus (Bovine) | SS |
| Q1RML2 | PLCZ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Bos taurus (Bovine) | PR |
| P10894 | PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Bos taurus (Bovine) | SS |
| Q2VRL0 | PLCZ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Gallus gallus (Chicken) | PR |
| P19174 | PLCG1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Homo sapiens (Human) | EV |
| Q8N3E9 | PLCD3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | Homo sapiens (Human) | SS |
| P16885 | PLCG2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 | Homo sapiens (Human) | SS |
| Q15111 | PLCL1 | Inactive phospholipase C-like protein 1 | Homo sapiens (Human) | PR |
| Q9UPR0 | PLCL2 | Inactive phospholipase C-like protein 2 | Homo sapiens (Human) | PR |
| Q86YW0 | PLCZ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Homo sapiens (Human) | PR |
| P51178 | PLCD1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Homo sapiens (Human) | EV |
| Q9BRC7 | PLCD4 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-4 | Homo sapiens (Human) | SS |
| Q00722 | PLCB2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | Homo sapiens (Human) | EV |
| Q01970 | PLCB3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Homo sapiens (Human) | EV |
| Q9NQ66 | PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Homo sapiens (Human) | EV |
| Q9P212 | PLCE1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Homo sapiens (Human) | SS |
| Q15147 | PLCB4 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-4 | Homo sapiens (Human) | PR |
| Q8K2J0 | Plcd3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | Mus musculus (Mouse) | PR |
| Q8K394 | Plcl2 | Inactive phospholipase C-like protein 2 | Mus musculus (Mouse) | PR |
| Q8K4S1 | Plce1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Mus musculus (Mouse) | SS |
| Q8R3B1 | Plcd1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Mus musculus (Mouse) | SS |
| P51432 | Plcb3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Mus musculus (Mouse) | SS |
| Q62077 | Plcg1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Mus musculus (Mouse) | SS |
| A3KGF7 | Plcb2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | Mus musculus (Mouse) | PR |
| Q9Z1B3 | Plcb1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Mus musculus (Mouse) | SS |
| Q7YRU3 | PLCZ | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Sus scrofa (Pig) | PR |
| P10686 | Plcg1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Rattus norvegicus (Rat) | EV |
| P24135 | Plcg2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2 | Rattus norvegicus (Rat) | SS |
| O89040 | Plcb2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | Rattus norvegicus (Rat) | PR |
| P10688 | Plcd1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Rattus norvegicus (Rat) | SS |
| P10687 | Plcb1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Rattus norvegicus (Rat) | SS |
| Q99P84 | Plce1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Rattus norvegicus (Rat) | EV |
| Q99JE6 | Plcb3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Rattus norvegicus (Rat) | SS |
| G5EBH0 | egl-8 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta egl-8 | Caenorhabditis elegans | SS |
| Q56W08 | PLC3 | Phosphoinositide phospholipase C 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NMA7 | PLC9 | Phosphoinositide phospholipase C 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GV43 | PLC6 | Phosphoinositide phospholipase C 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C2 | PLC5 | Phosphoinositide phospholipase C 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STZ3 | PLC8 | Phosphoinositide phospholipase C 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C1 | PLC4 | Phosphoinositide phospholipase C 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| A5D6R3 | plcd3a | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3-A | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKESFKVCFC | CVRNFKVKSS | EPPEEIKNLF | HDYSQDDRMS | ADEMLRFVIQ | VQGETHADIN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YVKDIFHRLK | HHGVFHPRGI | HLEGFYRYLL | SDFNSPLPLT | REVWQDMNQP | LSHYFLYTGH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NSYLTGNQLN | SNSSIEPIVK | ALRNGVRVIE | LDLWPNSSGK | EAEVRHGGTL | TSREDLQKCL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NVVKENAFQV | SAYPVVLTLE | DHLTPILQKK | VAKMVSKTFG | GSLFQCTDET | TECFPSPESL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KNKILISTKP | PKEYLQTQIS | KGSTTDESTR | AKKISDAEEQ | VQEEDEESVA | IEYRDLISIH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| AGNRKGGLKN | CLNGDPNRVI | RLSMSEQWLE | TLAKTRGPDL | VKFTQRNLLR | IFPKTTRFDS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SNYDPLVGWI | HGAQMVAFNM | QSHGRYLWMM | QGMFKANGGC | GYVKKPDVLL | SNGPEGEIFD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| PCSQNLPIKT | TLKVKIYTGE | GWNMDFPLDH | FDRYSPPDFY | AKVGIAGVPL | DTASYRTEID |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KDEWFPIWDK | EFEFPLRVPE | LSLLCITVKD | YDSNTQNDFA | GQTCFPLSEV | RPGIRAVRLH |
| 550 | 560 | ||||
| DRAGEVYKHV | RLLMRFVLEP | R |