Q38SD2
Gene name |
LRRK1 (KIAA1790) |
Protein name |
Leucine-rich repeat serine/threonine-protein kinase 1 |
Names |
EC 2.7.11.1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:79705 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
LEUCINE-RICH REPEAT RECEPTOR PROTEIN KINASE EMS1 (PTHR48055) |
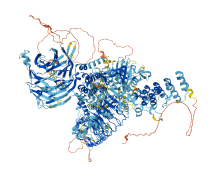
Descriptions
Leucine-rich repeat serine/threonine-protein kinase 1 (LRRK1) is a protein involved in the negative regulation of bone mass, acting through the maturation of osteoclasts. In contrast to the mechanism of autoinhibition that occurs in LRRK2, the autoinhibition of LRRK1 is mediated by a steric mechanism involving the ANK domain, which blocks access to the active site of the kinase domain in a dimer-dependent manner. In contrast, the interaction of the N-terminal LRR domain with the N-lobe of the kinase domain contributes to the alleviation of autoinhibition by changing its position relative to the WD40 domain, exposing the exposure of the active site of the kinase domain. The autoinhibited dimer of LRRK1 could be stabilized by a loop (1791-1907) in the WD40 domain. In addition, the kinase activity of LRRK1 is also regulated by the interaction of its COR-B loop with the kinase domain. Phosphorylation of three residues (S1064, T1074, and S1075) in the autoinhibitory COR-B loop significantly increases the kinase activity of LRRK1. Phosphorylation by Rab7a, resulting in the F1065A mutation, also alleviates the autoinhibited state of LRRK1.
Autoinhibitory domains (AIDs)
Target domain |
1263-1265 (N-lobe of kinase domain) |
Relief mechanism |
Partner binding, Cleavage |
Assay |
Structural analysis, Deletion assay |
Target domain |
1242-1525 (Kinase domain) |
Relief mechanism |
PTM, Partner binding, Cleavage |
Assay |
Structural analysis, Deletion assay |
Accessory elements
1408-1429 (Activation loop from InterPro)
Target domain |
1263-1265 (N-lobe of kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure
Activated structure
5 structures for Q38SD2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 8E04 | EM | 380 A | A | 20-2015 | PDB |
| 8E05 | EM | 460 A | A/B | 1-2015 | PDB |
| 8E06 | EM | 430 A | A | 1-2015 | PDB |
| 8FAC | EM | 392 A | A | 1-2015 | PDB |
| AF-Q38SD2-F1 | Predicted | AlphaFoldDB |
1993 variants for Q38SD2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV003365632 rs201543283 RCV001988892 |
4 | M>V | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002155620 rs200799740 RCV003025493 |
102 | M>I | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002008010 RCV003418273 RCV003264358 rs370235958 |
194 | V>M | LRRK1-related condition Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs368091026 RCV003339906 RCV002030943 |
215 | R>Q | Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [NCI-TCGA, ClinVar] | Yes |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV002548341 rs113989128 RCV000970015 |
224 | I>V | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002563640 rs145072406 RCV001942187 |
241 | I>L | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs749068051 RCV001886038 RCV002552206 |
287 | A>G | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
RCV002561542 rs372555242 RCV001974714 |
375 | N>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs199792081 RCV002556437 RCV001923485 |
586 | E>D | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001906414 rs150261606 RCV003264222 |
703 | P>L | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV002573552 COSM4053477 RCV002025042 COSM4053475 rs377308921 |
773 | T>M | Variant assessed as Somatic; MODERATE impact. Inborn genetic diseases [NCI-TCGA, ClinVar] | Yes |
NCI-TCGA Cosmic ClinVar 1000Genomes ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
RCV002561498 RCV001946388 rs202077818 |
829 | V>M | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs766875506 CA393966307 RCV000625749 |
929 | E>* | Osteosclerotic metaphyseal dysplasia [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
| VAR_086690 | 929 | E>del | OSMD [UniProt] | Yes | UniProt |
|
RCV002551070 rs185373027 RCV001870773 |
940 | S>C | Inborn genetic diseases [ClinVar] | Yes |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
RCV001887773 rs368618286 RCV002547910 |
1006 | M>V | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
RCV001925922 rs751191439 RCV003264250 |
1200 | R>Q | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs564175632 RCV003170195 RCV001968161 |
1202 | F>Y | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
rs1258583719 RCV002227891 |
1494 | R>* | Variant assessed as Somatic; HIGH impact. Osteosclerotic metaphyseal dysplasia [NCI-TCGA, ClinVar] | Yes |
ClinVar NCI-TCGA dbSNP gnomAD |
|
rs770946839 RCV002551626 RCV001877071 |
1760 | S>F | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV002489349 RCV000960070 rs34876840 |
1796 | P>H | Osteosclerotic metaphyseal dysplasia [ClinVar] | Yes |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
RCV002561510 RCV001955173 rs1301904566 |
1934 | E>D | Inborn genetic diseases [ClinVar] | Yes |
ClinVar TOPMed dbSNP gnomAD |
|
rs370701948 RCV002552935 RCV001871480 |
1945 | I>T | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs886038213 CA10586702 RCV001795385 |
1980 | E>missing | Osteosclerotic metaphyseal dysplasia [ClinVar] | Yes |
ClinGen ClinVar dbSNP |
|
RCV001794939 rs2141168796 |
1991 | A>missing | Osteosclerotic metaphyseal dysplasia [ClinVar] | Yes |
ClinVar dbSNP |
| rs781273894 | 2 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs779491648 | 4 | M>I | No |
ExAC gnomAD |
|
| rs201543283 | 4 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1191189390 | 4 | M>T | No | TOPMed | |
|
RCV002041255 rs748591648 |
5 | S>L | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs772698984 | 6 | Q>* | No |
ExAC gnomAD |
|
| rs1596149153 | 7 | R>S | No | Ensembl | |
| rs201657615 | 8 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs373263143 | 8 | P>S | No |
ESP TOPMed gnomAD |
|
| rs950488806 | 9 | P>L | No |
TOPMed gnomAD |
|
| rs950488806 | 9 | P>R | No |
TOPMed gnomAD |
|
| rs1438369630 | 10 | S>G | No |
TOPMed gnomAD |
|
| rs1422179063 | 10 | S>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1186222914 | 10 | S>R | No | gnomAD | |
| rs1429951456 | 11 | M>R | No |
TOPMed gnomAD |
|
| rs1429951456 | 11 | M>T | No |
TOPMed gnomAD |
|
| rs1366267504 | 11 | M>V | No |
TOPMed gnomAD |
|
| rs2042077612 | 12 | Y>C | No | gnomAD | |
| rs759385351 | 13 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs776094214 | 13 | W>L | No |
ExAC gnomAD |
|
| rs1228726751 | 14 | C>* | No |
TOPMed gnomAD |
|
|
RCV001938098 rs982310051 |
14 | C>S | No |
ClinVar TOPMed dbSNP gnomAD |
|
| TCGA novel | 14 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs866662547 | 14 | C>Y | No | Ensembl | |
| rs1304274213 | 15 | V>L | No | TOPMed | |
| rs1389883248 | 16 | G>V | No | gnomAD | |
| rs180759486 | 17 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3420175 rs1397828747 |
18 | E>D | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| rs1357600166 | 19 | E>A | No |
TOPMed gnomAD |
|
| rs1396018485 | 19 | E>K | No | gnomAD | |
| rs2042078545 | 21 | A>V | No | TOPMed | |
| rs1410381418 | 22 | V>G | No | TOPMed | |
| rs2042078587 | 22 | V>M | No | TOPMed | |
| rs2042078672 | 23 | C>Y | No | TOPMed | |
| rs1342350922 | 25 | E>Q | No | gnomAD | |
| rs767752687 | 26 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs756110175 | 26 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767752687 | 26 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs753949506 | 27 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs755378549 | 30 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs755378549 | 30 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs376424624 | 31 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs376424624 | 31 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2042079260 | 32 | N>D | No | gnomAD | |
| rs1596149347 | 32 | N>S | No | Ensembl | |
| rs2141653359 | 33 | G>A | No | Ensembl | |
| rs778209303 | 33 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1054254534 | 34 | A>V | No | TOPMed | |
| rs1011877404 | 35 | G>A | No |
TOPMed gnomAD |
|
| rs1011877404 | 35 | G>E | No |
TOPMed gnomAD |
|
| rs1442417412 | 36 | D>Y | No | gnomAD | |
| rs1018256609 | 37 | T>M | No |
TOPMed gnomAD |
|
| rs1377840329 | 38 | G>D | No |
TOPMed gnomAD |
|
| rs1377840329 | 38 | G>V | No |
TOPMed gnomAD |
|
| rs1386565028 | 39 | G>C | No |
TOPMed gnomAD |
|
| rs1455503071 | 39 | G>D | No | gnomAD | |
| rs1386565028 | 39 | G>R | No |
TOPMed gnomAD |
|
| rs1386565028 | 39 | G>S | No |
TOPMed gnomAD |
|
| rs753089141 | 41 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs753089141 | 41 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs1296797976 | 41 | P>S | No | TOPMed | |
| rs1346090652 | 42 | S>F | No | TOPMed | |
| rs2031116479 | 44 | R>P | No | TOPMed | |
|
TCGA novel rs2031116479 |
44 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1385978407 | 45 | G>V | No | gnomAD | |
| rs2031118023 | 46 | G>D | No | TOPMed | |
| rs2031117780 | 46 | G>S | No |
TOPMed gnomAD |
|
| rs998158742 | 47 | D>N | No | TOPMed | |
| rs2031118961 | 48 | P>L | No | TOPMed | |
| rs777607829 | 48 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs777607829 | 48 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs954117551 | 49 | A>P | No |
TOPMed gnomAD |
|
| rs954117551 | 49 | A>S | No |
TOPMed gnomAD |
|
| rs954117551 | 49 | A>T | No |
TOPMed gnomAD |
|
| rs1007479203 | 50 | A>E | No |
TOPMed gnomAD |
|
| rs1007479203 | 50 | A>V | No |
TOPMed gnomAD |
|
| rs2031119712 | 51 | R>L | No | TOPMed | |
| rs1360870166 | 52 | S>A | No |
TOPMed gnomAD |
|
| rs1293448832 | 52 | S>C | No |
TOPMed gnomAD |
|
| rs1314649839 | 53 | R>H | No |
TOPMed gnomAD |
|
| rs756857060 | 54 | R>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs564533545 RCV001971862 |
54 | R>W | No |
ClinVar 1000Genomes TOPMed dbSNP gnomAD |
|
| rs2031122033 | 55 | T>M | No | gnomAD | |
| rs2031122479 | 57 | G>A | No | TOPMed | |
| TCGA novel | 57 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs941012164 | 59 | R>C | No |
TOPMed gnomAD |
|
|
RCV001981813 rs780551753 |
59 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2031123612 | 61 | A>S | No |
TOPMed gnomAD |
|
| rs1350030008 | 61 | A>V | No | gnomAD | |
| rs918823575 | 62 | Y>C | No |
TOPMed gnomAD |
|
| rs918823575 | 62 | Y>F | No |
TOPMed gnomAD |
|
|
rs915826670 RCV001910187 |
63 | R>K | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs915826670 | 63 | R>T | No |
TOPMed gnomAD |
|
| rs2031124641 | 64 | R>G | No | Ensembl | |
|
rs948639431 RCV001900062 |
64 | R>L | No |
ClinVar TOPMed dbSNP |
|
| rs948639431 | 64 | R>P | No | TOPMed | |
| rs1189245756 | 65 | G>E | No |
TOPMed gnomAD |
|
| rs2031125255 | 65 | G>R | No | TOPMed | |
| rs1281309106 | 66 | D>E | No |
TOPMed gnomAD |
|
| rs1481018763 | 66 | D>N | No |
TOPMed gnomAD |
|
| rs1481018763 | 66 | D>Y | No |
TOPMed gnomAD |
|
|
RCV002076432 rs528714330 |
67 | R>C | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs528714330 | 67 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2031126507 | 67 | R>H | No | Ensembl | |
| rs1054604781 | 68 | G>A | No | TOPMed | |
|
RCV001985236 rs56289455 |
68 | G>C | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
RCV002125034 rs56289455 |
68 | G>S | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs2031127555 | 69 | G>S | No | Ensembl | |
| rs914448297 | 70 | A>S | No |
TOPMed gnomAD |
|
| rs914448297 | 70 | A>T | No |
TOPMed gnomAD |
|
| rs934768991 | 71 | R>G | No |
TOPMed gnomAD |
|
| rs1326828605 | 71 | R>P | No | TOPMed | |
| rs934768991 | 71 | R>W | No |
TOPMed gnomAD |
|
| rs1443319958 | 72 | D>E | No |
TOPMed gnomAD |
|
| rs2141653682 | 75 | E>Q | No | Ensembl | |
| rs2031130166 | 76 | E>A | No | TOPMed | |
| rs945844784 | 76 | E>D | No | TOPMed | |
| rs1053194090 | 76 | E>K | No | TOPMed | |
|
RCV001981333 rs893166977 |
77 | A>G | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs768515004 | 77 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs893166977 | 77 | A>V | No |
TOPMed gnomAD |
|
|
RCV001864924 rs1249255322 |
79 | D>G | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1456154770 | 79 | D>N | No |
TOPMed gnomAD |
|
| rs1456154770 | 79 | D>Y | No |
TOPMed gnomAD |
|
| rs562434357 | 80 | Q>* | No |
1000Genomes TOPMed gnomAD |
|
| rs562434357 | 80 | Q>E | No |
1000Genomes TOPMed gnomAD |
|
| rs562434357 | 80 | Q>K | No |
1000Genomes TOPMed gnomAD |
|
| rs1340750957 | 81 | C>Y | No |
TOPMed gnomAD |
|
| rs761654387 | 84 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs796076773 | 85 | L>M | No |
TOPMed gnomAD |
|
| rs997739606 | 86 | E>K | No | gnomAD | |
|
RCV001971132 rs28468535 |
87 | K>E | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs758349602 | 87 | K>T | No | Ensembl | |
| rs755637633 | 88 | G>S | No |
ExAC gnomAD |
|
|
RCV001997059 rs779865899 |
90 | L>P | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs779865899 | 90 | L>R | No |
ExAC gnomAD |
|
| rs2031713947 | 90 | L>V | No | gnomAD | |
| rs370948444 | 94 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs202037702 | 94 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed |
| rs901489770 | 95 | A>T | No |
TOPMed gnomAD |
|
| rs377500835 | 96 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs377500835 | 96 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs773149239 | 97 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1309717731 | 97 | Y>H | No | gnomAD | |
| rs2031715971 | 100 | L>P | No | TOPMed | |
| rs1460054859 | 101 | E>D | No |
TOPMed gnomAD |
|
|
rs775753659 RCV002040397 |
104 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs775753659 | 104 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1427176646 | 104 | R>H | No |
TOPMed gnomAD |
|
|
TCGA novel rs1427176646 |
104 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1449166266 | 105 | Y>* | No | gnomAD | |
| rs1596234842 | 105 | Y>S | No | Ensembl | |
| rs1567214660 | 106 | L>V | No | Ensembl | |
| rs1187686666 | 107 | L>F | No | gnomAD | |
|
rs202052348 RCV002021726 |
108 | S>N | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs575177605 | 110 | R>K | No |
1000Genomes ExAC gnomAD |
|
| rs774548299 | 110 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2031718367 | 112 | V>M | No | TOPMed | |
| rs2031718507 | 114 | L>V | No | TOPMed | |
| rs2031718645 | 115 | P>S | No | Ensembl | |
| rs951902777 | 116 | T>I | No | gnomAD | |
| rs1467463832 | 116 | T>P | No | Ensembl | |
| rs2031719556 | 117 | E>K | No | Ensembl | |
| rs750884820 | 119 | T>M | No | ExAC | |
| rs1383049081 | 120 | D>E | No | gnomAD | |
| rs1185218356 | 121 | D>A | No | TOPMed | |
| rs1433919455 | 121 | D>H | No | gnomAD | |
| rs1433919455 | 121 | D>N | No | gnomAD | |
| rs1364561496 | 122 | N>D | No | gnomAD | |
| rs1364561496 | 122 | N>H | No | gnomAD | |
| rs1215618264 | 122 | N>I | No | gnomAD | |
| rs1215618264 | 122 | N>S | No | gnomAD | |
| rs1215618264 | 122 | N>T | No | gnomAD | |
| rs753606461 | 125 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs753606461 RCV001872926 |
125 | V>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs754657738 | 128 | A>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 128 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs754657738 COSM5848860 COSM5848858 |
128 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs776974790 | 129 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs758151687 | 130 | F>L | No |
ExAC gnomAD |
|
| rs200045067 | 133 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2031723466 | 136 | V>L | No | TOPMed | |
| rs201198678 | 137 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2031723958 | 138 | E>K | No |
TOPMed gnomAD |
|
|
COSM3815663 COSM4923959 rs775524685 COSM4923961 COSM3815665 |
139 | L>F | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs374355989 | 140 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3886696 COSM3886694 |
142 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1260718403 | 146 | P>L | No |
TOPMed gnomAD |
|
| rs762328731 | 146 | P>S | No |
ExAC gnomAD |
|
| rs763758358 | 147 | C>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs536752090 | 148 | S>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs536752090 | 148 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1261409516 | 148 | S>R | No | gnomAD | |
| rs1436826237 | 149 | P>L | No | TOPMed | |
|
COSM5277250 COSM5277252 |
151 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756058323 | 151 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM3386775 COSM3386777 rs552211004 |
151 | R>W | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1400336345 | 153 | L>R | No |
TOPMed gnomAD |
|
| rs1174295697 | 154 | N>I | No | gnomAD | |
| rs1412103908 | 157 | L>Q | No |
TOPMed gnomAD |
|
|
rs2141669802 RCV001887459 |
159 | L>F | No |
ClinVar Ensembl dbSNP |
|
| rs752855869 | 163 | R>* | No |
ExAC TOPMed gnomAD |
|
|
RCV001889550 rs200548454 |
163 | R>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs778159326 | 164 | G>V | No |
ExAC gnomAD |
|
|
COSM4053457 COSM4053455 |
168 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1457392995 | 168 | V>F | No | TOPMed | |
| rs1457392995 | 168 | V>I | No | TOPMed | |
| rs2032002385 | 169 | V>L | No | TOPMed | |
| rs1032895584 | 171 | L>F | No |
TOPMed gnomAD |
|
| rs1032895584 | 171 | L>I | No |
TOPMed gnomAD |
|
|
rs202112893 COSM199319 |
175 | T>M | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs2032003446 | 176 | H>R | No | gnomAD | |
| rs1207247731 | 177 | G>E | No | gnomAD | |
| rs1484726939 | 177 | G>R | No |
TOPMed gnomAD |
|
| TCGA novel | 178 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs554667646 | 180 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs767351188 | 181 | E>A | No |
ExAC gnomAD |
|
| rs761442314 | 181 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs773030697 | 182 | S>R | No |
ExAC gnomAD |
|
| rs757738305 | 183 | Y>C | No |
ExAC gnomAD |
|
|
rs753664394 COSM959603 |
184 | A>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs919254235 | 185 | V>F | No | gnomAD | |
| rs1404204859 | 185 | V>G | No | gnomAD | |
| rs919254235 | 185 | V>L | No | gnomAD | |
| rs751875924 | 186 | R>G | No |
ExAC gnomAD |
|
| rs757519760 | 186 | R>S | No |
ExAC gnomAD |
|
| rs1357929316 | 187 | K>R | No |
TOPMed gnomAD |
|
| rs2032006656 | 189 | E>A | No | TOPMed | |
| rs930941488 | 189 | E>D | No |
TOPMed gnomAD |
|
| rs746264958 | 189 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1596241797 | 190 | F>Y | No | Ensembl | |
| rs756374275 | 193 | I>N | No |
ExAC gnomAD |
|
| rs370235958 | 194 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs201889926 COSM959605 COSM2200425 |
195 | R>C | Variant assessed as Somatic; MODERATE impact. central_nervous_system endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
COSM959607 rs773830700 |
195 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs201889926 | 195 | R>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2032008547 | 198 | L>V | No | Ensembl | |
| rs1884063698 | 199 | Y>* | No | gnomAD | |
| rs747840699 | 200 | A>S | No |
ExAC gnomAD |
|
| rs747840699 | 200 | A>T | No |
ExAC gnomAD |
|
| rs771745497 | 200 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs766026615 | 201 | A>T | No |
ExAC gnomAD |
|
| rs1161513298 | 202 | I>M | No | gnomAD | |
|
rs35985016 RCV002167046 |
203 | K>E | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1458359895 | 203 | K>M | No | gnomAD | |
| TCGA novel | 205 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759520031 | 205 | G>R | No |
ExAC gnomAD |
|
| rs769939723 | 206 | N>H | No | Ensembl | |
| rs769871825 | 208 | D>N | No |
ExAC gnomAD |
|
| rs1205832502 | 211 | I>M | No | gnomAD | |
| rs2032034285 | 212 | F>L | No |
TOPMed gnomAD |
|
| rs557184600 | 215 | R>G | No |
1000Genomes ExAC gnomAD |
|
| rs368091026 | 215 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs557184600 | 215 | R>W | No |
1000Genomes ExAC gnomAD |
|
| rs1417277255 | 217 | G>R | No | gnomAD | |
| TCGA novel | 217 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1596242577 TCGA novel |
218 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs767489943 | 219 | Y>S | No |
ExAC gnomAD |
|
| rs1428019785 | 221 | C>S | No |
TOPMed gnomAD |
|
| rs1428019785 | 221 | C>Y | No |
TOPMed gnomAD |
|
| rs1253523506 | 222 | S>C | No |
TOPMed gnomAD |
|
| rs1398112465 | 222 | S>P | No | gnomAD | |
| rs2032036272 | 223 | Y>C | No | TOPMed | |
| rs2032036603 | 226 | L>V | No | TOPMed | |
| rs766808583 | 228 | S>N | No |
ExAC gnomAD |
|
| rs1359433750 | 228 | S>R | No | gnomAD | |
|
COSM4053461 COSM4053463 |
229 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754234882 | 230 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs754234882 | 230 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs755289273 | 231 | P>A | No |
ExAC gnomAD |
|
| rs2141670682 | 231 | P>L | No | Ensembl | |
| rs755289273 | 231 | P>S | No |
ExAC gnomAD |
|
|
rs755289273 RCV001943528 |
231 | P>T | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1321549084 | 238 | K>N | No | gnomAD | |
| rs1282669597 | 238 | K>R | No | gnomAD | |
| rs1282669597 | 238 | K>T | No | gnomAD | |
| rs145072406 | 241 | I>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs558815397 | 241 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs145072406 | 241 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201348332 | 243 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs780846312 | 245 | P>T | No |
ExAC gnomAD |
|
| rs1489194935 | 246 | L>S | No | gnomAD | |
| rs924383338 | 247 | P>H | No |
TOPMed gnomAD |
|
| rs745664601 | 248 | S>G | No |
ExAC gnomAD |
|
| rs1428568069 | 248 | S>T | No | gnomAD | |
| rs369239695 | 251 | P>A | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs200690035 | 251 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs369239695 | 251 | P>S | No |
1000Genomes ESP TOPMed gnomAD |
|
|
COSM3987866 COSM3987868 |
254 | T>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1437413884 | 254 | T>S | No |
TOPMed gnomAD |
|
| rs2033103774 | 257 | R>C | No | TOPMed | |
| rs1324210109 | 257 | R>H | No | TOPMed | |
| rs747061264 | 259 | K>E | No |
ExAC gnomAD |
|
|
rs770982171 COSM4406082 COSM4406080 |
261 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs776892472 | 262 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs369330842 | 263 | L>H | No |
ESP ExAC TOPMed |
|
| rs765643116 | 268 | V>E | No |
ExAC gnomAD |
|
| rs763339239 | 269 | D>A | No |
ExAC gnomAD |
|
| rs1439992221 | 269 | D>E | No |
TOPMed gnomAD |
|
| rs2033105932 | 269 | D>H | No |
TOPMed gnomAD |
|
| rs752048498 | 274 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs756776320 | 275 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs199863201 | 276 | I>S | No |
1000Genomes ExAC |
|
| rs1164314445 | 278 | C>R | No | gnomAD | |
| rs750140074 | 279 | Q>H | No |
ExAC gnomAD |
|
| rs1408267488 | 280 | I>V | No |
TOPMed gnomAD |
|
| rs1333528063 | 281 | T>M | No |
TOPMed gnomAD |
|
| rs2033107521 | 282 | E>K | No | gnomAD | |
| TCGA novel | 283 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs755805466 | 283 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs1429167452 | 284 | D>H | No | TOPMed | |
| rs1380348674 | 286 | S>F | No | gnomAD | |
|
rs779802602 RCV001881899 |
286 | S>P | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs2141691592 RCV001970614 |
287 | A>P | No |
ClinVar Ensembl dbSNP |
|
| rs749068051 | 287 | A>V | No |
ExAC gnomAD |
|
| rs754733732 | 288 | N>K | No |
ExAC gnomAD |
|
| rs1046335520 | 291 | A>E | No |
TOPMed gnomAD |
|
| rs2033108674 | 291 | A>T | No | gnomAD | |
| rs1046335520 | 291 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 292 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033109024 | 292 | T>I | No | TOPMed | |
| rs373434869 | 293 | L>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs373434869 | 293 | L>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1259431781 | 293 | L>V | No |
TOPMed gnomAD |
|
| rs1202661217 | 294 | P>S | No | gnomAD | |
| rs1398231033 | 296 | V>A | No |
TOPMed gnomAD |
|
| rs1334256380 | 296 | V>I | No | gnomAD | |
| rs1188757802 | 298 | P>L | No | gnomAD | |
|
COSM3499719 COSM3499717 |
298 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776656740 | 303 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs745902377 | 304 | L>V | No |
ExAC gnomAD |
|
| rs201297016 | 305 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs775625158 | 305 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1404679405 | 306 | K>N | No | gnomAD | |
| rs2141691698 | 307 | L>V | No | Ensembl | |
| rs1277823709 | 308 | N>Y | No | TOPMed | |
| TCGA novel | 310 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769053443 | 311 | D>N | No |
ExAC gnomAD |
|
| rs762350334 | 313 | H>L | No |
ExAC TOPMed gnomAD |
|
| rs762350334 | 313 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs762350334 | 313 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1270878753 | 314 | L>V | No |
TOPMed gnomAD |
|
| rs766132517 | 315 | G>R | No |
1000Genomes ExAC gnomAD |
|
| rs574940704 | 316 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs574940704 | 316 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778760210 | 317 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1405888383 | 318 | P>L | No | TOPMed | |
| rs1445730152 | 319 | G>D | No |
TOPMed gnomAD |
|
|
RCV002004310 rs2141691789 |
319 | G>H | No |
ClinVar Ensembl dbSNP |
|
| rs1261488928 | 319 | G>R | No |
TOPMed gnomAD |
|
| rs1261488928 | 319 | G>S | No |
TOPMed gnomAD |
|
| rs376445041 | 320 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs376445041 | 320 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1480137858 | 321 | Q>P | No | gnomAD | |
| rs2141691813 | 322 | S>T | No | Ensembl | |
| rs769809859 | 323 | S>* | No |
ExAC TOPMed gnomAD |
|
| rs1195530604 | 323 | S>P | No | Ensembl | |
|
RCV000880312 rs184008590 |
324 | D>E | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
TCGA novel rs2033115026 |
325 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs2033115270 | 327 | I>F | No | gnomAD | |
| rs1267255810 | 331 | L>P | No | gnomAD | |
| rs770384623 | 332 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1296621966 | 333 | E>K | No |
TOPMed gnomAD |
|
| rs562018856 | 334 | I>T | No |
1000Genomes ExAC gnomAD |
|
| rs2033176484 | 334 | I>V | No | Ensembl | |
| rs1441786750 | 336 | I>V | No | gnomAD | |
| TCGA novel | 337 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1308229623 | 338 | S>G | No |
TOPMed gnomAD |
|
|
rs368549166 RCV001977304 |
338 | S>R | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1162564377 | 340 | K>N | No | gnomAD | |
| rs1443887829 | 340 | K>R | No | gnomAD | |
| rs1351595378 | 341 | L>F | No |
TOPMed gnomAD |
|
| rs1490853664 | 342 | S>P | No |
TOPMed gnomAD |
|
| TCGA novel | 344 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1447370113 | 344 | L>V | No | TOPMed | |
| rs1397593141 | 345 | P>R | No | gnomAD | |
| rs866765967 | 345 | P>S | No | gnomAD | |
| rs866765967 | 345 | P>T | No | gnomAD | |
| rs1448546235 | 346 | P>A | No | gnomAD | |
| rs1448546235 | 346 | P>S | No | gnomAD | |
| rs187360115 | 347 | G>E | No | 1000Genomes | |
|
TCGA novel rs1310385882 |
348 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1418693007 | 349 | L>W | No | Ensembl | |
| rs762797401 | 350 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs763873193 | 351 | L>F | No |
ExAC gnomAD |
|
| rs751540811 | 352 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs2033179466 | 354 | L>V | No | gnomAD | |
| rs150351407 | 355 | Q>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2141693546 | 356 | K>Q | No | Ensembl | |
| rs1596268585 | 357 | L>R | No | Ensembl | |
| rs767309864 | 358 | T>R | No |
ExAC gnomAD |
|
| rs1459224772 | 359 | A>T | No |
TOPMed gnomAD |
|
| rs1199543291 | 360 | S>T | No | gnomAD | |
|
COSM2200438 COSM2200440 |
362 | N>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs753981637 | 363 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs1482604307 | 365 | E>Q | No | Ensembl | |
| rs779206703 | 366 | K>Q | No |
ExAC gnomAD |
|
| rs1424330941 | 367 | L>* | No | gnomAD | |
|
rs748365815 RCV001933945 |
367 | L>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs372847198 | 369 | E>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs372847198 RCV002130506 COSM1375687 |
369 | E>K | large_intestine [Cosmic] | No |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs368813810 | 373 | A>V | No |
ESP ExAC gnomAD |
|
| rs566606745 | 374 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs566606745 | 374 | T>P | No |
1000Genomes ExAC gnomAD |
|
| rs566606745 | 374 | T>S | No |
1000Genomes ExAC gnomAD |
|
| rs765224234 | 376 | W>R | No |
ExAC gnomAD |
|
| rs1489302353 | 377 | I>T | No | TOPMed | |
|
rs200469257 RCV001874078 |
378 | G>A | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs200469257 | 378 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1243857037 | 379 | L>S | No |
TOPMed gnomAD |
|
| rs778089814 | 380 | R>Q | No |
ExAC gnomAD |
|
| rs758667510 | 380 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2033191949 | 382 | L>V | No |
TOPMed gnomAD |
|
|
rs2141693825 RCV001946453 |
384 | E>* | No |
ClinVar Ensembl dbSNP |
|
| rs1229067054 | 385 | L>I | No | TOPMed | |
| rs547363177 | 386 | D>V | No |
ExAC gnomAD |
|
| rs1489266948 | 387 | I>L | No | gnomAD | |
| rs757510968 | 387 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1193689209 | 387 | I>T | No |
TOPMed gnomAD |
|
|
rs1476817125 RCV001919958 |
390 | N>S | No |
ClinVar TOPMed dbSNP gnomAD |
|
|
RCV001933952 rs371086173 |
391 | K>I | No |
ClinVar ESP ExAC dbSNP gnomAD |
|
| rs769386932 | 393 | T>A | No |
ExAC gnomAD |
|
| rs1291141402 | 393 | T>I | No | gnomAD | |
| rs769386932 | 393 | T>S | No |
ExAC gnomAD |
|
| rs2141693881 | 394 | E>* | No | Ensembl | |
| rs2033194969 | 395 | L>P | No | TOPMed | |
| rs1391841773 | 396 | P>R | No | TOPMed | |
|
COSM2200453 COSM2200451 |
396 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200253977 | 397 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200253977 | 397 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1420851289 COSM959613 |
397 | A>V | endometrium [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs768329041 | 398 | L>V | No |
ExAC gnomAD |
|
| rs549021266 | 399 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs761682123 | 400 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs771748188 | 401 | H>Y | No |
ExAC gnomAD |
|
| rs1306810748 | 402 | S>P | No |
TOPMed gnomAD |
|
| rs2033196586 | 405 | S>A | No | Ensembl | |
| rs2033196715 | 405 | S>F | No | Ensembl | |
| rs773266521 | 406 | L>F | No |
ExAC gnomAD |
|
| rs1217292614 | 407 | N>D | No | gnomAD | |
| rs760495714 | 407 | N>S | No |
ExAC TOPMed gnomAD |
|
| COSM959615 | 408 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375638857 | 412 | S>C | No |
TOPMed gnomAD |
|
| rs375638857 | 412 | S>F | No |
TOPMed gnomAD |
|
|
RCV000892937 rs55739947 |
416 | L>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1218547521 | 416 | L>Q | No | gnomAD | |
| rs1177097335 | 418 | V>A | No | gnomAD | |
| rs763083060 | 418 | V>L | No |
ExAC TOPMed gnomAD |
|
|
RCV001944801 rs538150906 |
420 | P>L | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1209278460 | 426 | P>A | No | TOPMed | |
| rs1349991935 | 426 | P>R | No |
TOPMed gnomAD |
|
| TCGA novel | 427 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs767732473 | 428 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs767732473 | 428 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs1453654500 | 429 | C>R | No |
TOPMed gnomAD |
|
| rs201450091 | 429 | C>Y | No | 1000Genomes | |
| TCGA novel | 430 | C>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1318234747 | 430 | C>F | No | gnomAD | |
| rs2033276081 | 436 | A>V | No | TOPMed | |
| TCGA novel | 437 | L>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1188820322 | 438 | E>G | No | gnomAD | |
| rs1476877345 | 439 | C>F | No | gnomAD | |
| rs1476877345 | 439 | C>Y | No | gnomAD | |
| rs1317836448 | 441 | P>A | No | TOPMed | |
| rs754548120 | 442 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2033277373 | 442 | D>N | No | TOPMed | |
|
rs1015865458 RCV002012762 |
442 | D>V | No |
ClinVar TOPMed dbSNP gnomAD |
|
| TCGA novel | 444 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM5159291 COSM1375689 |
445 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033278238 | 445 | A>S | No | gnomAD | |
| rs747881165 | 445 | A>V | No |
ExAC gnomAD |
|
| rs758006341 | 446 | V>A | No |
ExAC gnomAD |
|
| rs758006341 | 446 | V>G | No |
ExAC gnomAD |
|
| rs1174654799 | 446 | V>I | No |
TOPMed gnomAD |
|
| rs993194467 | 447 | F>L | No |
TOPMed gnomAD |
|
| rs76152020 | 448 | W>L | No | Ensembl | |
| rs2033279349 | 449 | K>E | No |
TOPMed gnomAD |
|
| rs2033279468 | 450 | N>S | No | Ensembl | |
| rs1157264053 | 451 | H>L | No | Ensembl | |
|
RCV001974536 rs777438481 |
452 | L>P | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2033280122 | 453 | K>* | No | TOPMed | |
|
COSM6141652 COSM6141654 |
453 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1486538796 | 453 | K>R | No |
TOPMed gnomAD |
|
| rs1487100650 | 454 | D>E | No | TOPMed | |
| rs200529094 | 454 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200529094 | 454 | D>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2033280662 | 456 | D>G | No | TOPMed | |
| COSM282559 | 457 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346889073 | 459 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 460 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs113678526 RCV000947652 |
461 | A>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs531721861 | 462 | L>F | No |
1000Genomes ExAC gnomAD |
|
| rs768909431 | 463 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1487597371 | 463 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2033281795 | 464 | E>Q | No | Ensembl | |
| rs371606687 | 466 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1264026847 COSM4699232 COSM4699234 |
466 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| TCGA novel | 467 | L>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1269637042 | 467 | L>R | No |
TOPMed gnomAD |
|
| rs1478879427 | 470 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1174095366 | 473 | D>A | No | gnomAD | |
| rs1174095366 | 473 | D>V | No | gnomAD | |
| TCGA novel | 474 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033426302 | 475 | L>H | No | Ensembl | |
| rs1321714856 | 476 | M>K | No | gnomAD | |
| rs772177705 | 476 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs772177705 | 476 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs773371877 | 477 | F>L | No |
ExAC gnomAD |
|
| rs1488899950 | 479 | R>T | No |
TOPMed gnomAD |
|
| rs1198284104 | 481 | Q>R | No |
TOPMed gnomAD |
|
| rs1456270070 | 483 | N>Y | No | gnomAD | |
| rs1251076782 | 485 | L>V | No | gnomAD | |
| rs377571378 | 486 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1232533272 | 487 | A>S | No |
TOPMed gnomAD |
|
| rs1323910825 | 490 | P>S | No |
TOPMed gnomAD |
|
| rs753186926 | 491 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1169485935 | 494 | W>R | No |
TOPMed gnomAD |
|
| rs2033428973 | 497 | R>T | No | TOPMed | |
| rs2033429191 | 499 | L>I | No | Ensembl | |
| rs757916117 | 501 | T>I | No |
ExAC gnomAD |
|
| rs1394823786 | 504 | L>R | No | gnomAD | |
| rs2033430021 | 510 | G>A | No | Ensembl | |
| rs566734053 | 511 | K>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
RCV001945546 rs770937949 |
511 | K>N | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs1335513063 | 511 | K>R | No | gnomAD | |
| rs1270102762 | 512 | N>S | No | TOPMed | |
|
COSM5212675 COSM1477913 |
513 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1049141464 | 513 | E>K | No |
TOPMed gnomAD |
|
| rs2141700870 | 514 | D>Y | No | Ensembl | |
| rs770108501 | 516 | L>P | No |
ExAC gnomAD |
|
| rs2033481892 | 517 | K>E | No | gnomAD | |
| rs1365834896 | 518 | T>M | No |
TOPMed gnomAD |
|
| rs1567233858 | 519 | K>N | No | Ensembl | |
|
RCV001981986 rs774941330 |
520 | R>C | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs2033482549 | 520 | R>H | No | Ensembl | |
|
COSM3969001 COSM3969003 |
521 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs868044422 | 521 | I>V | No | Ensembl | |
| rs767129254 | 523 | F>V | No |
ExAC gnomAD |
|
| rs375733293 | 524 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1267137737 | 526 | T>I | No |
TOPMed gnomAD |
|
| rs777091251 | 527 | R>G | No |
TOPMed gnomAD |
|
| rs1226232670 | 528 | G>D | No | TOPMed | |
| rs1245703001 | 529 | R>C | No |
TOPMed gnomAD |
|
| rs765898071 | 529 | R>L | No |
ExAC gnomAD |
|
| rs754659010 | 530 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs200499483 | 531 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001983596 rs200499483 |
531 | R>G | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2033484669 | 531 | R>H | No | Ensembl | |
| rs2033484883 | 532 | S>P | No | Ensembl | |
| rs781229505 | 533 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs781229505 | 533 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs780075028 | 534 | T>I | No |
ExAC gnomAD |
|
| rs780075028 | 534 | T>S | No |
ExAC gnomAD |
|
| rs1013772472 | 535 | E>D | No |
TOPMed gnomAD |
|
| rs1327622975 | 536 | A>T | No | gnomAD | |
| rs1464285760 | 537 | A>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2033486005 | 537 | A>P | No | TOPMed | |
| rs2141708186 | 538 | S>C | No | Ensembl | |
| rs904019203 | 541 | E>K | No |
TOPMed gnomAD |
|
| rs772736703 | 543 | P>L | No |
ExAC TOPMed gnomAD |
|
|
rs55798315 RCV000968486 |
543 | P>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs763899406 | 544 | A>T | No |
TOPMed gnomAD |
|
| rs2033760973 | 545 | F>S | No |
TOPMed gnomAD |
|
| rs2033761084 | 546 | L>P | No | Ensembl | |
| TCGA novel | 547 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1296485632 | 548 | E>V | No |
TOPMed gnomAD |
|
| rs1459531837 | 549 | S>P | No |
TOPMed gnomAD |
|
| rs776409174 | 552 | V>F | No |
ExAC gnomAD |
|
| rs776409174 | 552 | V>I | No |
ExAC gnomAD |
|
| rs759169632 | 553 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1423124755 | 554 | C>Y | No | TOPMed | |
| rs769390447 | 555 | L>V | No |
ExAC gnomAD |
|
| rs762674468 | 557 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs764032483 | 560 | L>I | No |
ExAC gnomAD |
|
| rs764032483 | 560 | L>V | No |
ExAC gnomAD |
|
| rs761695009 | 561 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs761695009 | 561 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1198339219 | 562 | T>I | No |
TOPMed gnomAD |
|
| rs766339820 | 563 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs766339820 | 563 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1262313834 | 564 | P>L | No | TOPMed | |
| rs867186599 | 564 | P>S | No |
TOPMed gnomAD |
|
|
RCV001985306 rs867186599 |
564 | P>T | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs141522582 | 566 | S>L | No |
1000Genomes ExAC gnomAD |
|
| rs1373918258 | 567 | V>I | No | gnomAD | |
| rs753116269 | 568 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs1304804237 | 568 | C>R | No | TOPMed | |
| TCGA novel | 570 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1403388422 | 571 | K>E | No | TOPMed | |
| rs2033765412 | 572 | S>G | No | TOPMed | |
| rs758773787 | 572 | S>N | No | ExAC | |
|
COSM4826930 COSM4826932 |
574 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1026190245 | 576 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1026190245 | 576 | L>I | No | gnomAD | |
| rs780592294 | 576 | L>P | No |
ExAC gnomAD |
|
| rs1319866402 | 577 | Y>C | No | gnomAD | |
| rs1319866402 | 577 | Y>F | No | gnomAD | |
| rs2033766607 | 578 | L>M | No | gnomAD | |
| rs745344763 | 579 | G>V | No |
ExAC gnomAD |
|
| rs779750560 | 582 | P>A | No |
ExAC gnomAD |
|
| rs1270375539 | 584 | L>V | No |
TOPMed gnomAD |
|
|
COSM6141649 COSM6141651 |
585 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs538821865 | 585 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs538821865 RCV002032100 |
585 | R>Q | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs768546791 | 585 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1275278452 | 587 | L>V | No | gnomAD | |
| rs771907397 | 588 | P>L | No |
ExAC gnomAD |
|
| rs771907397 | 588 | P>R | No |
ExAC gnomAD |
|
| rs759743498 | 589 | P>H | No |
ExAC gnomAD |
|
| rs759743498 | 589 | P>R | No |
ExAC gnomAD |
|
| rs773225777 | 589 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs2033812975 | 593 | Q>L | No | Ensembl | |
| rs1251231353 | 595 | G>S | No | gnomAD | |
| rs2033813542 | 596 | N>S | No | TOPMed | |
|
RCV001900833 rs2033813786 |
597 | L>F | No |
ClinVar TOPMed dbSNP |
|
| rs1455216844 | 598 | W>* | No | Ensembl | |
| rs1455216844 | 598 | W>S | No | Ensembl | |
| rs2033814151 | 599 | Q>* | No | TOPMed | |
| rs2033814151 | 599 | Q>K | No | TOPMed | |
| rs1419500207 | 600 | L>P | No | gnomAD | |
|
RCV001910933 rs763061068 |
602 | T>I | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs536343284 | 604 | D>E | No |
1000Genomes ExAC gnomAD |
|
|
COSM4053464 COSM4053466 |
605 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033814795 | 606 | T>A | No | TOPMed | |
| rs376893013 | 609 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4925081 COSM4925079 |
611 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1299227526 | 612 | A>T | No |
TOPMed gnomAD |
|
| rs767970862 | 614 | I>T | No |
ExAC gnomAD |
|
| rs1412937940 | 615 | Q>E | No | gnomAD | |
| rs2141709839 | 615 | Q>R | No | Ensembl | |
| rs755662621 | 617 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs80186667 | 618 | G>A | No | Ensembl | |
| COSM959617 | 618 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1389322413 | 619 | P>L | No | TOPMed | |
| rs2033836110 | 620 | K>R | No |
TOPMed gnomAD |
|
| rs753290789 | 621 | A>V | No |
ExAC gnomAD |
|
| rs754452197 | 622 | M>T | No |
ExAC gnomAD |
|
| rs1471655500 | 624 | S>F | No |
TOPMed gnomAD |
|
| rs200770615 | 624 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM2200499 COSM2200498 rs747828168 |
627 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001895518 rs753941048 |
627 | R>H | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs753941048 | 627 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1245381002 | 628 | A>G | No | TOPMed | |
|
rs1176065275 COSM5350241 COSM5350242 |
629 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
|
rs1006607984 RCV001892954 |
630 | L>R | No |
ClinVar TOPMed dbSNP gnomAD |
|
|
rs779184003 COSM1213957 |
631 | R>Q | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
RCV002048456 rs192470343 |
631 | R>W | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1347137222 | 632 | K>Q | No |
TOPMed gnomAD |
|
| rs749390141 | 633 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2033839853 | 635 | K>M | No | Ensembl | |
| rs1596286684 | 635 | K>Q | No | TOPMed | |
| rs762074642 | 636 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs762074642 | 636 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1305528910 | 637 | K>R | No | gnomAD | |
| rs1320984844 | 638 | L>Q | No | Ensembl | |
| rs772234195 | 639 | M>I | No |
ExAC gnomAD |
|
|
COSM699631 COSM1646651 |
640 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2033840823 | 641 | M>I | No | TOPMed | |
| rs2033840973 | 642 | I>N | No | Ensembl | |
| rs918407051 | 643 | I>L | No | TOPMed | |
| rs918407051 | 643 | I>V | No | TOPMed | |
|
COSM1375691 COSM5158930 rs375903897 |
644 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2033841599 | 645 | G>S | No | TOPMed | |
| rs377316707 | 646 | P>S | No |
ESP ExAC gnomAD |
|
| rs377316707 | 646 | P>T | No |
ESP ExAC gnomAD |
|
| rs764768964 | 647 | P>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 647 | P>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3499729 COSM3499727 |
647 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs200971195 | 648 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs777502572 | 648 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 648 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777502572 | 648 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1188594611 | 649 | Q>H | No |
TOPMed gnomAD |
|
| rs2033842452 | 650 | G>S | No | TOPMed | |
| rs949321749 | 653 | T>A | No | Ensembl | |
| rs2033842901 | 653 | T>I | No | Ensembl | |
| rs34885781 | 654 | L>I | No | gnomAD | |
| rs2033843754 | 657 | I>L | No | TOPMed | |
| rs781346039 | 657 | I>M | No |
ExAC gnomAD |
|
| rs530260600 | 660 | T>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4475303 RCV001919286 rs530260600 COSM4475302 |
660 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar 1000Genomes ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs1392637520 | 661 | G>E | No |
TOPMed gnomAD |
|
| rs1164323692 | 661 | G>W | No |
TOPMed gnomAD |
|
| rs779074952 | 662 | R>K | No |
ExAC gnomAD |
|
| rs2033844987 | 662 | R>S | No | TOPMed | |
| rs2033845095 | 663 | A>V | No | TOPMed | |
| rs748140184 | 664 | P>R | No |
ExAC gnomAD |
|
| rs1276075553 | 665 | Q>E | No | TOPMed | |
| TCGA novel | 665 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs773362599 | 666 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs773362599 | 666 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2033846366 | 668 | H>L | No | gnomAD | |
| rs1332242404 | 668 | H>Y | No |
TOPMed gnomAD |
|
|
COSM4148340 rs2141710821 |
669 | G>* | thyroid [Cosmic] | No |
cosmic curated Ensembl |
| rs777044065 | 669 | G>A | No |
ExAC gnomAD |
|
| rs935196455 | 670 | E>D | No | TOPMed | |
| TCGA novel | 670 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
RCV001958290 rs745630986 |
672 | T>missing | No |
ClinVar dbSNP |
|
| rs760065482 | 672 | T>A | No |
ExAC gnomAD |
|
| rs760065482 | 672 | T>P | No |
ExAC gnomAD |
|
| rs765611843 | 676 | T>A | No |
ExAC gnomAD |
|
| rs2141710869 | 677 | K>R | No | Ensembl | |
| rs1235154576 | 678 | W>G | No | gnomAD | |
| rs1415877583 | 683 | P>L | No |
TOPMed gnomAD |
|
| rs1166776778 | 684 | A>V | No | gnomAD | |
| TCGA novel | 687 | R>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3499730 COSM3499732 |
690 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs919588747 | 690 | V>F | No | Ensembl | |
| rs755804910 | 691 | E>G | No |
ExAC gnomAD |
|
| TCGA novel | 691 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs2033946833 RCV001864587 |
691 | E>Q | No |
ClinVar Ensembl dbSNP |
|
|
RCV001876479 rs200409352 |
692 | S>T | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs201243082 | 693 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1567241312 | 694 | E>G | No | Ensembl | |
| rs76435780 | 696 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs141102052 RCV000963337 |
697 | V>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1344347761 | 700 | I>V | No | TOPMed | |
| rs896004560 | 701 | G>R | No |
TOPMed gnomAD |
|
| rs770051407 | 702 | G>A | No |
ExAC TOPMed gnomAD |
|
|
COSM3886699 COSM3886697 |
702 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1393261925 | 704 | A>G | No | gnomAD | |
| rs769240723 | 705 | S>I | No |
ExAC gnomAD |
|
| rs774033253 | 706 | M>I | No |
ExAC gnomAD |
|
| rs761343909 | 707 | A>T | No |
ExAC gnomAD |
|
| TCGA novel | 710 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1287711282 | 711 | Q>K | No | gnomAD | |
| rs1259266503 | 713 | F>C | No | TOPMed | |
| rs767215413 | 713 | F>V | No |
ExAC gnomAD |
|
| rs2033950214 | 714 | F>L | No | gnomAD | |
| rs78191111 | 715 | T>R | No | Ensembl | |
| rs766053466 | 716 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs760582606 | 716 | D>G | No |
ExAC gnomAD |
|
| rs758131541 | 716 | D>Y | No | Ensembl | |
| rs753818870 | 718 | A>T | No |
ExAC gnomAD |
|
| rs1328893943 | 721 | V>A | No | TOPMed | |
| rs199919214 | 721 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV002047822 rs199919214 |
721 | V>M | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1285293554 | 724 | W>* | No | gnomAD | |
| COSM1323737 | 725 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1297141389 | 726 | L>V | No | TOPMed | |
|
COSM4053469 COSM4053471 rs891430665 |
727 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1455548471 | 730 | E>G | No | gnomAD | |
| rs1198900798 | 731 | E>K | No | gnomAD | |
| rs1198900798 | 731 | E>Q | No | gnomAD | |
| rs2033953814 | 732 | A>V | No |
TOPMed gnomAD |
|
| rs370663048 | 733 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs996103849 | 735 | N>H | No | Ensembl | |
| rs2033954489 | 735 | N>S | No | TOPMed | |
|
rs562726580 RCV002120540 |
736 | L>V | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1461201082 | 738 | F>L | No | gnomAD | |
| rs746593625 | 741 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2033955141 | 741 | L>H | No | gnomAD | |
| rs769293742 | 742 | N>S | No |
ExAC gnomAD |
|
| rs2033955564 | 743 | I>N | No | TOPMed | |
| rs371661382 | 744 | E>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs371661382 | 744 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1567242360 | 746 | K>M | No | Ensembl | |
| rs2034009123 | 747 | A>T | No | TOPMed | |
| rs1371799804 | 749 | N>D | No |
TOPMed gnomAD |
|
| rs772262731 | 750 | A>T | No |
ExAC TOPMed gnomAD |
|
|
RCV001914906 rs141579296 |
751 | V>M | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs887555486 | 753 | L>P | No | TOPMed | |
| rs760948329 | 756 | G>R | No | Ensembl | |
| rs1172054475 | 757 | T>M | No | TOPMed | |
|
COSM433601 COSM5200933 |
758 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1014001464 | 759 | L>R | No | Ensembl | |
| rs1230489888 | 759 | L>V | No |
TOPMed gnomAD |
|
| rs2034011945 | 760 | D>N | No | TOPMed | |
| rs199817137 | 761 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs896287484 | 762 | I>F | No |
TOPMed gnomAD |
|
| rs765670323 | 762 | I>T | No |
ExAC gnomAD |
|
| rs2034012746 | 763 | E>K | No | TOPMed | |
| rs2034013235 | 765 | K>N | No | TOPMed | |
| rs1200186227 | 765 | K>R | No |
TOPMed gnomAD |
|
| rs1200186227 | 765 | K>T | No |
TOPMed gnomAD |
|
| rs1186256525 | 767 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs753053998 COSM3815674 COSM3815672 |
767 | R>H | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs753053998 | 767 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs2034014085 | 770 | R>K | No | Ensembl | |
|
RCV001892357 rs201803929 |
771 | I>T | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs373497333 | 772 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 774 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1018773132 | 775 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs368099995 | 775 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs368099995 | 775 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs768495809 | 778 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs768495809 | 778 | V>E | No |
ExAC TOPMed gnomAD |
|
| rs2034016684 | 778 | V>M | No | TOPMed | |
| rs1231380055 | 780 | A>T | No | gnomAD | |
| rs2141101055 | 780 | A>V | No | Ensembl | |
| rs572305968 | 783 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777040315 | 783 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs759855609 | 784 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs1258652050 | 784 | S>Y | No |
TOPMed gnomAD |
|
| rs1466528093 | 785 | P>H | No | gnomAD | |
| rs1252856565 | 786 | S>C | No | gnomAD | |
| rs1252856565 | 786 | S>F | No | gnomAD | |
|
rs367783860 COSM959624 |
787 | G>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs752076949 | 788 | S>P | No |
ExAC gnomAD |
|
| TCGA novel | 790 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1567242706 | 791 | T>A | No |
TOPMed gnomAD |
|
| rs1167430688 | 791 | T>I | No | gnomAD | |
|
COSM3886700 COSM3886702 |
792 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2034020368 | 797 | T>A | No | gnomAD | |
| rs756811495 | 797 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2034020634 | 798 | F>C | No | Ensembl | |
| rs2141101125 | 798 | F>L | No | Ensembl | |
| rs2034020759 | 799 | K>T | No | Ensembl | |
| rs780703616 | 800 | H>N | No |
ExAC gnomAD |
|
| rs745342762 | 800 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs780703616 | 800 | H>Y | No |
ExAC gnomAD |
|
| rs2141102369 | 802 | H>Q | No | Ensembl | |
| rs1276599427 | 802 | H>Y | No | gnomAD | |
| rs1040913056 | 803 | E>G | No | Ensembl | |
| rs1040913056 | 803 | E>V | No | Ensembl | |
|
COSM2200542 COSM959626 |
805 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2034073805 | 809 | L>P | No | gnomAD | |
| rs372434815 | 810 | E>K | No |
ESP ExAC gnomAD |
|
| rs866015235 | 811 | G>S | No | Ensembl | |
| rs200114885 | 814 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774702868 | 814 | G>V | No |
ExAC gnomAD |
|
|
rs766532218 COSM4053480 COSM4053478 |
816 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs762111517 | 816 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs762111517 | 816 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs762111517 | 816 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs761403767 | 817 | Q>H | No | gnomAD | |
| rs2034076259 | 819 | I>T | No | Ensembl | |
| rs1213306218 | 819 | I>V | No | gnomAD | |
| rs750927387 | 820 | F>L | No |
ExAC gnomAD |
|
| rs1263333149 | 821 | H>L | No |
TOPMed gnomAD |
|
| rs760146211 | 821 | H>Y | No |
ExAC gnomAD |
|
|
COSM3386780 rs1050845680 COSM3386778 |
822 | V>I | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1050845680 | 822 | V>L | No |
TOPMed gnomAD |
|
| rs941918894 | 826 | M>L | No | Ensembl | |
| rs778637944 | 826 | M>T | No |
ExAC gnomAD |
|
| rs2034078064 | 827 | K>E | No | Ensembl | |
| rs373879296 | 827 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 828 | D>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1596294295 | 828 | D>A | No | Ensembl | |
| rs1399654915 | 829 | V>A | No |
TOPMed gnomAD |
|
| rs200452644 | 830 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs186890317 | 831 | S>N | No | 1000Genomes | |
| rs1415786642 | 832 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1596294339 | 832 | T>P | No | Ensembl | |
| rs370572047 | 833 | I>N | No |
ESP TOPMed |
|
| rs749260427 | 833 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs768937081 | 834 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1478755633 | 835 | C>S | No | TOPMed | |
| rs1596294380 | 835 | C>W | No | Ensembl | |
| rs1366577662 | 837 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs774547176 | 837 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2034081222 | 838 | L>M | No | TOPMed | |
| rs1007554321 | 841 | R>Q | No |
TOPMed gnomAD |
|
| rs375374079 | 841 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1329603545 | 844 | P>H | No | gnomAD | |
| rs759150062 | 845 | R>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 845 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1596294939 | 847 | Y>S | No | Ensembl | |
| rs775330321 | 848 | L>M | No |
ExAC TOPMed gnomAD |
|
| rs775330321 | 848 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs762727247 | 849 | S>R | No |
ExAC TOPMed |
|
|
rs763773289 COSM98988 |
852 | E>K | stomach [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs751385962 | 853 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1023999423 | 854 | V>A | No | Ensembl | |
| rs368404584 | 854 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
rs368404584 RCV001991418 |
854 | V>M | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| TCGA novel | 859 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750298507 | 860 | R>C | No |
ExAC TOPMed gnomAD |
|
|
RCV001988799 COSM959628 rs142470911 |
860 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ClinVar 1000Genomes ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs981947042 | 861 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs778989938 | 861 | R>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 863 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777797735 | 863 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs758619910 | 863 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs915301795 | 864 | D>H | No |
1000Genomes TOPMed gnomAD |
|
| rs915301795 | 864 | D>N | No |
1000Genomes TOPMed gnomAD |
|
| rs202089769 | 865 | D>H | No |
ExAC TOPMed gnomAD |
|
|
rs202089769 RCV002125828 |
865 | D>N | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
RCV002173199 rs56003881 |
866 | D>N | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs374008052 | 867 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374008052 | 867 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1274852106 | 868 | Q>* | No | gnomAD | |
| rs560090431 | 868 | Q>L | No |
TOPMed gnomAD |
|
| rs560090431 | 868 | Q>R | No |
TOPMed gnomAD |
|
| rs1179791606 | 869 | Y>C | No | gnomAD | |
| rs1258675580 | 870 | L>P | No | gnomAD | |
| rs762496843 | 871 | T>M | No |
ExAC gnomAD |
|
| rs773874591 | 872 | D>G | No |
ExAC gnomAD |
|
| rs2034105491 | 872 | D>Y | No | Ensembl | |
| rs1169332520 | 876 | E>* | No | gnomAD | |
| rs1169332520 | 876 | E>K | No | gnomAD | |
| rs1169332520 | 876 | E>Q | No | gnomAD | |
| rs761710931 | 877 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs1464705980 | 879 | V>G | No | gnomAD | |
| rs1222722240 | 879 | V>M | No | TOPMed | |
|
COSM5209538 COSM433603 |
880 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs531640499 | 880 | E>D | No |
1000Genomes ExAC gnomAD |
|
| rs767199367 | 880 | E>G | No |
ExAC gnomAD |
|
| rs1304862195 | 880 | E>K | No | gnomAD | |
| rs1304862195 | 880 | E>Q | No | gnomAD | |
| rs767199367 | 880 | E>V | No |
ExAC gnomAD |
|
| rs1296705143 | 881 | Q>H | No | gnomAD | |
| rs201854079 | 882 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752784877 | 883 | P>S | No |
ExAC gnomAD |
|
| rs758457191 | 884 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs758457191 | 884 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs997852963 | 886 | D>N | No |
TOPMed gnomAD |
|
| rs757689685 | 887 | I>M | No |
ExAC gnomAD |
|
| rs77112930 | 888 | K>R | No | Ensembl | |
| rs2141103392 | 889 | D>E | No | Ensembl | |
| rs1189823477 | 889 | D>G | No | gnomAD | |
| rs1451916944 | 889 | D>N | No | gnomAD | |
| rs866448925 | 890 | Y>H | No | Ensembl | |
| rs746169733 | 891 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2034110305 | 893 | L>V | No | TOPMed | |
| rs1466315314 | 895 | S>L | No | gnomAD | |
| rs747624120 | 896 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs200667677 | 898 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200667677 | 898 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2034161280 | 899 | F>C | No | gnomAD | |
| rs1247341695 | 900 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2141104648 | 901 | I>M | No | Ensembl | |
| rs1224886642 | 901 | I>T | No | TOPMed | |
| rs1596296664 | 903 | T>P | No | Ensembl | |
| rs1482022169 | 904 | G>D | No | gnomAD | |
| rs2034162012 | 904 | G>S | No | Ensembl | |
| rs1596296692 | 905 | T>P | No | Ensembl | |
| rs1180277045 | 905 | T>S | No |
TOPMed gnomAD |
|
| rs1412669248 | 907 | L>I | No | gnomAD | |
| rs770680533 | 907 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1596296729 | 909 | F>S | No | Ensembl | |
| rs1050786363 | 910 | P>L | No | gnomAD | |
| rs765181449 | 912 | T>P | No |
ExAC gnomAD |
|
| rs774309946 | 915 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1003990525 | 920 | Y>C | No | Ensembl | |
| rs2034164808 | 921 | F>L | No | Ensembl | |
| rs2034164929 | 922 | L>R | No |
TOPMed gnomAD |
|
| rs200062338 | 923 | D>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200062338 | 923 | D>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM959632 | 926 | W>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766875506 | 929 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs766875506 | 929 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs2034165777 | 930 | C>Y | No |
TOPMed gnomAD |
|
| rs1596296819 | 933 | R>M | No | Ensembl | |
| TCGA novel | 936 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1182311678 | 939 | G>C | No | Ensembl | |
| rs754220038 | 939 | G>D | No |
ExAC gnomAD |
|
| rs185373027 | 940 | S>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs755449019 | 940 | S>P | No |
ExAC gnomAD |
|
| rs757918696 | 941 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs747666136 RCV001902644 |
941 | R>W | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2034167443 | 946 | N>K | No | Ensembl | |
| rs2034167749 | 948 | V>M | No | TOPMed | |
| TCGA novel | 949 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1183522126 | 950 | R>T | No | gnomAD | |
| rs1596296895 | 951 | A>T | No | Ensembl | |
| rs746639649 | 951 | A>V | No |
ExAC gnomAD |
|
| rs190712554 | 953 | D>E | No |
1000Genomes ExAC gnomAD |
|
| rs201392064 | 955 | R>K | No |
1000Genomes ExAC gnomAD |
|
| rs769677938 | 955 | R>S | No |
ExAC gnomAD |
|
| rs775494533 | 956 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs1349039201 | 958 | L>Q | No | TOPMed | |
| rs1285701374 | 959 | V>G | No | TOPMed | |
| rs988874365 | 959 | V>L | No | Ensembl | |
| rs761840209 | 961 | T>I | No |
ExAC gnomAD |
|
| rs761840209 | 961 | T>N | No |
ExAC gnomAD |
|
| rs796226537 | 964 | T>E | No | Ensembl | |
| rs767685227 | 964 | T>M | No |
ExAC TOPMed gnomAD |
|
|
COSM4053484 COSM4053486 |
965 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1304950127 | 966 | Q>L | No | Ensembl | |
| rs1343031369 | 967 | T>A | No |
TOPMed gnomAD |
|
|
rs41531245 RCV002082476 |
967 | T>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs369170053 | 968 | E>K | No | gnomAD | |
| rs2034172256 | 969 | E>Q | No |
TOPMed gnomAD |
|
| rs947265550 | 971 | Y>C | No |
TOPMed gnomAD |
|
| rs374599598 | 972 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1178431017 | 972 | F>L | No |
TOPMed gnomAD |
|
|
RCV002036677 rs201547686 |
974 | F>L | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2034172925 | 974 | F>Y | No | TOPMed | |
| rs559037479 | 976 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs559037479 | 976 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2034173400 | 976 | A>V | No | TOPMed | |
|
COSM2200584 COSM2200582 rs939724696 |
981 | A>T | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes NCI-TCGA TOPMed gnomAD |
| rs2141104979 | 983 | P>A | No | Ensembl | |
|
COSM2200585 COSM2200587 rs369265377 |
984 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2034174505 | 985 | A>T | No |
TOPMed gnomAD |
|
| rs1323525873 | 986 | N>H | No | TOPMed | |
|
RCV002047743 rs2141105006 |
986 | N>K | No |
ClinVar Ensembl dbSNP |
|
|
RCV002033030 rs199709127 |
986 | N>S | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs377315974 | 987 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs768678229 | 988 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs768678229 | 988 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1184817709 | 992 | P>L | No | gnomAD | |
|
rs2141122311 RCV002008826 |
994 | L>V | No |
ClinVar Ensembl dbSNP |
|
|
COSM1646649 COSM699627 |
997 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1364842626 | 997 | S>P | No | gnomAD | |
| rs2035055800 | 998 | K>E | No | TOPMed | |
| rs2035055947 | 998 | K>I | No |
TOPMed gnomAD |
|
| rs1457821862 | 1000 | G>D | No | gnomAD | |
| rs2035056520 | 1002 | D>N | No | TOPMed | |
| rs769865724 | 1002 | D>V | No | ExAC | |
| rs764652283 | 1004 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs2035057482 | 1005 | G>D | No | TOPMed | |
| rs761362971 | 1005 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs2035057868 | 1006 | M>T | No | Ensembl | |
| rs367709807 | 1007 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs370459525 COSM4053487 COSM4053489 |
1007 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1272354949 | 1008 | H>Y | No |
TOPMed gnomAD |
|
| rs1436285093 | 1009 | P>S | No |
TOPMed gnomAD |
|
| rs2035059099 | 1013 | T>A | No | Ensembl | |
| rs754811151 | 1015 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs2035059498 | 1016 | R>W | No | Ensembl | |
| rs1596316921 | 1017 | V>E | No | Ensembl | |
| TCGA novel | 1019 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1176301432 | 1019 | K>T | No |
TOPMed gnomAD |
|
| rs1251670485 | 1020 | M>I | No | gnomAD | |
| rs2035060003 | 1020 | M>R | No | TOPMed | |
| rs2035060418 | 1022 | F>L | No |
TOPMed gnomAD |
|
| rs781362754 | 1023 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2035060707 | 1024 | P>A | No |
TOPMed gnomAD |
|
| rs770068206 | 1025 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs368567214 | 1026 | G>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1299019662 | 1032 | I>K | No | gnomAD | |
|
rs1247301494 RCV001902806 |
1032 | I>M | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1299019662 | 1032 | I>T | No | gnomAD | |
| rs749693795 | 1034 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs372293144 | 1034 | R>W | No |
ESP TOPMed gnomAD |
|
| rs1245145585 | 1035 | M>I | No | gnomAD | |
| rs768987575 | 1035 | M>T | No |
ExAC gnomAD |
|
| rs538312065 | 1038 | S>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs538312065 | 1038 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs766117523 | 1040 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2035063823 | 1045 | Q>L | No | TOPMed | |
| rs1265360267 | 1048 | E>G | No | gnomAD | |
| rs1208647497 | 1054 | K>E | No | gnomAD | |
| rs1246733997 | 1055 | S>G | No | gnomAD | |
|
COSM1300959 rs748456823 |
1058 | R>K | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2035214274 | 1059 | K>N | No |
TOPMed gnomAD |
|
| rs2035214454 | 1061 | T>P | No | Ensembl | |
| rs1351909744 | 1062 | I>L | No |
TOPMed gnomAD |
|
| rs1351909744 | 1062 | I>V | No |
TOPMed gnomAD |
|
|
COSM4053490 COSM4053492 |
1066 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1067 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1178570157 | 1068 | N>K | No | gnomAD | |
| rs2035215310 | 1070 | R>G | No | Ensembl | |
| rs1431147853 | 1070 | R>K | No | gnomAD | |
| COSM261855 | 1071 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs376281560 | 1072 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs765787797 | 1072 | R>H | No |
ExAC gnomAD |
|
| rs776235920 | 1073 | C>Y | No |
ExAC gnomAD |
|
| rs2035216151 | 1074 | S>G | No | gnomAD | |
| rs759296759 | 1074 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs2035216378 | 1076 | F>L | No | gnomAD | |
| rs2035216496 | 1077 | R>* | No | TOPMed | |
| rs2035216783 | 1078 | V>M | No | Ensembl | |
| rs1018354863 | 1082 | Q>H | No |
TOPMed gnomAD |
|
| rs1304931060 | 1083 | T>A | No |
TOPMed gnomAD |
|
|
rs539691388 RCV002161480 |
1084 | I>T | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2035217377 | 1085 | Y>D | No | TOPMed | |
| rs1393239281 | 1086 | W>C | No |
TOPMed gnomAD |
|
| rs2035217511 | 1087 | Q>* | No | Ensembl | |
| rs1596320974 | 1088 | E>G | No | Ensembl | |
|
COSM2200620 rs752554538 COSM2200618 |
1088 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1321235742 | 1090 | L>F | No |
TOPMed gnomAD |
|
| rs1287460559 | 1090 | L>H | No | gnomAD | |
| rs1321235742 | 1090 | L>V | No |
TOPMed gnomAD |
|
| rs1596321002 | 1092 | V>G | No | Ensembl | |
| rs1324636253 | 1096 | G>E | No | gnomAD | |
| rs1266729329 | 1097 | G>C | No | gnomAD | |
| rs1319288561 | 1097 | G>D | No | gnomAD | |
| rs1266729329 | 1097 | G>S | No | gnomAD | |
| rs200404538 | 1099 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200404538 | 1099 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1428684279 | 1102 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2035287557 | 1102 | E>D | No | TOPMed | |
| rs371351101 | 1105 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV002078397 rs78587240 |
1105 | D>N | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs200111263 | 1106 | V>L | No | gnomAD | |
| rs200111263 | 1106 | V>M | No | gnomAD | |
| rs1055979271 | 1107 | N>Y | No | TOPMed | |
| rs1396900032 | 1109 | K>Q | No | TOPMed | |
|
RCV001980349 rs2141128456 |
1112 | K>Q | No |
ClinVar Ensembl dbSNP |
|
| rs2035289511 | 1113 | S>N | No | TOPMed | |
| rs762543442 | 1113 | S>R | No |
ExAC TOPMed gnomAD |
|
|
rs768286875 COSM271369 |
1114 | G>R | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2035290072 | 1115 | G>E | No | Ensembl | |
| rs41425846 | 1118 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs2035290715 | 1120 | C>W | No | Ensembl | |
|
COSM3420178 COSM3420176 |
1121 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773055907 | 1121 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1459904257 | 1124 | V>E | No |
TOPMed gnomAD |
|
|
RCV001883045 rs373841325 |
1125 | R>G | No |
ClinVar ESP TOPMed dbSNP gnomAD |
|
| rs2035291515 | 1126 | D>E | No | Ensembl | |
|
COSM4816556 COSM4816554 |
1128 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2035291823 | 1129 | A>D | No | Ensembl | |
| rs2035291659 | 1129 | A>P | No | TOPMed | |
| rs2035292134 | 1131 | A>V | No | gnomAD | |
| rs765327562 | 1134 | T>A | No |
ExAC gnomAD |
|
| rs61733312 | 1134 | T>K | No |
ExAC TOPMed gnomAD |
|
|
COSM255292 rs61733312 |
1134 | T>M | central_nervous_system [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs61733312 | 1134 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1455038791 | 1135 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs764322028 | 1135 | D>Y | No |
ExAC gnomAD |
|
| rs1401599388 | 1137 | V>A | No |
TOPMed gnomAD |
|
| rs537781752 | 1137 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1202181508 | 1139 | S>A | No |
TOPMed gnomAD |
|
| TCGA novel | 1139 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3499736 COSM3499738 |
1142 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1143 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2035295313 | 1143 | Q>P | No | Ensembl | |
| TCGA novel | 1147 | A>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376554922 | 1147 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs376554922 | 1147 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs747816549 | 1149 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs1596326746 | 1149 | T>P | No | Ensembl | |
| rs2035455837 | 1151 | T>I | No | TOPMed | |
| rs2035456492 | 1154 | D>G | No | TOPMed | |
| rs1290143171 | 1154 | D>N | No |
TOPMed gnomAD |
|
| rs762983452 | 1155 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs768759171 | 1156 | T>M | No |
ExAC gnomAD |
|
| rs765489207 | 1157 | P>L | No | Ensembl | |
| rs1283656818 | 1158 | L>V | No | gnomAD | |
| rs767789230 | 1159 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1470869746 | 1161 | Q>E | No | Ensembl | |
| rs1281214969 | 1162 | Y>C | No | gnomAD | |
| rs761113327 | 1163 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs761113327 | 1163 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2141132820 | 1164 | P>S | No | Ensembl | |
| rs1222833998 | 1167 | V>I | No | gnomAD | |
| rs2141132873 | 1169 | E>G | No | Ensembl | |
| rs755387791 | 1169 | E>K | No |
ExAC gnomAD |
|
| rs755387791 | 1169 | E>Q | No |
ExAC gnomAD |
|
| rs1472484324 | 1174 | Q>P | No | gnomAD | |
| rs1161712901 | 1175 | H>D | No | gnomAD | |
| rs369305577 | 1176 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1307730712 | 1177 | D>E | No |
TOPMed gnomAD |
|
|
COSM4922915 COSM4922913 |
1177 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758062446 | 1178 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs758062446 | 1178 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs960673143 | 1178 | P>S | No | Ensembl | |
| rs960673143 | 1178 | P>T | No | Ensembl | |
| rs777394343 | 1179 | S>N | No |
ExAC gnomAD |
|
| rs777394343 | 1179 | S>T | No |
ExAC gnomAD |
|
| rs374717787 | 1181 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2035461405 | 1182 | S>P | No | TOPMed | |
| rs1245205004 | 1183 | E>K | No |
TOPMed gnomAD |
|
| rs2035462081 | 1184 | D>N | No | TOPMed | |
| rs200666835 | 1185 | V>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200666835 | 1185 | V>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs866854209 | 1185 | V>M | No | gnomAD | |
|
RCV001938185 rs373432119 |
1186 | Q>R | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM699625 COSM1646648 |
1187 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774585715 | 1189 | D>A | No |
ExAC gnomAD |
|
| rs769551074 | 1189 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs769551074 | 1189 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs532404694 | 1190 | M>L | No |
1000Genomes ExAC gnomAD |
|
| rs1182348413 | 1190 | M>T | No |
TOPMed gnomAD |
|
| rs532404694 | 1190 | M>V | No |
1000Genomes ExAC gnomAD |
|
| rs1367751693 | 1191 | E>D | No | Ensembl | |
| rs773533129 | 1191 | E>K | No |
ExAC gnomAD |
|
| rs760991902 | 1192 | D>E | No |
ExAC gnomAD |
|
| rs1460242604 | 1195 | L>R | No | gnomAD | |
| rs754107420 | 1196 | T>M | No |
ExAC TOPMed gnomAD |
|
|
COSM3886703 COSM3886705 |
1197 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs369473298 | 1198 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV002011624 rs1050305838 |
1198 | I>V | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs566023115 | 1199 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs777443874 | 1200 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs267604401 | 1201 | D>N | No | Ensembl | |
| rs2035470096 | 1203 | I>M | No |
TOPMed gnomAD |
|
| TCGA novel | 1207 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781044673 | 1207 | R>K | No |
ExAC gnomAD |
|
|
COSM3987869 COSM3987871 |
1208 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1596327227 | 1208 | H>P | No | TOPMed | |
| rs1596327227 | 1208 | H>R | No | TOPMed | |
| rs769826948 | 1209 | P>A | No |
ExAC gnomAD |
|
|
rs774546820 COSM3401592 COSM3401594 |
1209 | P>L | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs2141133179 RCV001970134 |
1211 | L>missing | No |
ClinVar dbSNP |
|
| rs2035472094 | 1211 | L>F | No | Ensembl | |
| rs773373328 | 1212 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs773373328 | 1212 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs772254004 | 1212 | P>S | No |
ExAC gnomAD |
|
| rs2035473432 | 1213 | V>A | No | TOPMed | |
| rs199624259 | 1213 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs199624259 RCV001889966 |
1213 | V>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs370568105 | 1214 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1216 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775945942 | 1216 | Q>P | No |
ExAC gnomAD |
|
| rs2035474283 | 1217 | E>D | No | gnomAD | |
|
COSM6141646 COSM6141648 |
1217 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2035474433 | 1221 | E>K | No | Ensembl | |
|
RCV001927570 rs751030086 |
1226 | D>N | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2035475124 | 1227 | F>L | No | TOPMed | |
| rs370451042 | 1228 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA gnomAD |
| rs2035475693 | 1229 | A>P | No | Ensembl | |
| rs2035551848 | 1232 | F>C | No | TOPMed | |
| rs1296908321 | 1234 | E>D | No |
TOPMed gnomAD |
|
| rs2035552288 | 1234 | E>K | No | TOPMed | |
| rs2035552288 | 1234 | E>Q | No | TOPMed | |
| rs1376209745 | 1235 | N>D | No |
TOPMed gnomAD |
|
| rs375762978 | 1236 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs370150056 | 1237 | K>N | No |
ESP ExAC gnomAD |
|
|
rs373500031 RCV002043765 |
1239 | E>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1429110266 | 1240 | H>D | No |
TOPMed gnomAD |
|
| rs1391459204 | 1240 | H>Q | No | gnomAD | |
|
RCV001870506 rs1429110266 |
1240 | H>Y | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs780403145 | 1241 | S>I | No |
ExAC gnomAD |
|
| rs377311326 | 1241 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs547256891 | 1242 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1596329517 | 1243 | D>N | No | Ensembl | |
| rs558530256 | 1244 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs558530256 | 1244 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1048503724 | 1245 | G>S | No | Ensembl | |
| rs772735483 | 1246 | S>N | No |
ExAC gnomAD |
|
| rs2035558359 | 1247 | V>A | No | gnomAD | |
| rs200086296 | 1247 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM6076724 COSM6076722 |
1249 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1567271146 | 1250 | Q>E | No | Ensembl | |
| rs1567271167 | 1251 | G>S | No | Ensembl | |
| rs373729202 | 1252 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs758309073 | 1256 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
|
COSM98989 rs201939674 |
1257 | I>V | stomach [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs1596329644 | 1258 | Y>S | No | Ensembl | |
| rs200606953 | 1259 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs200606953 RCV002088502 |
1259 | R>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs750451092 | 1259 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2035561045 | 1260 | A>S | No | TOPMed | |
|
RCV002023332 rs199933427 |
1261 | R>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs780250032 | 1261 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2035561515 | 1263 | Q>* | No | TOPMed | |
| rs2035561665 | 1264 | G>S | No | TOPMed | |
| rs1338947308 | 1266 | P>S | No | gnomAD | |
| rs966424843 | 1267 | V>M | No | Ensembl | |
| rs2035562257 | 1268 | A>V | No | Ensembl | |
| rs779284426 | 1269 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2035562719 | 1270 | K>E | No | gnomAD | |
| rs748630672 | 1271 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs748630672 | 1271 | R>G | No |
ExAC TOPMed gnomAD |
|
|
rs199804667 COSM959634 RCV001922161 |
1271 | R>H | endometrium [Cosmic] | No |
cosmic curated ClinVar ESP ExAC TOPMed dbSNP gnomAD |
| rs199804667 | 1271 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs748630672 | 1271 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs911429879 | 1272 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1184937563 | 1277 | F>C | No | gnomAD | |
|
rs778455063 COSM4699246 COSM4699244 |
1277 | F>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs1413148326 | 1277 | F>L | No | TOPMed | |
| rs1487742727 | 1278 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs893772249 | 1279 | N>K | No |
TOPMed gnomAD |
|
| rs2035565605 | 1281 | A>S | No | gnomAD | |
| rs1474867399 | 1281 | A>V | No | gnomAD | |
| rs539324349 | 1282 | N>K | No |
1000Genomes ExAC gnomAD |
|
| rs752497934 | 1283 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs752497934 | 1283 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1466480542 | 1284 | P>A | No | gnomAD | |
| rs145412332 | 1284 | P>L | No |
1000Genomes TOPMed gnomAD |
|
| rs145412332 | 1284 | P>Q | No |
1000Genomes TOPMed gnomAD |
|
|
COSM4995548 COSM4995550 |
1287 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1433263726 | 1287 | T>S | No |
TOPMed gnomAD |
|
| rs765481948 | 1288 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2035580324 | 1289 | L>P | No | Ensembl | |
| rs1301601156 | 1290 | R>K | No | gnomAD | |
| rs1363417994 | 1291 | H>N | No | gnomAD | |
| rs895020670 | 1292 | L>M | No | Ensembl | |
| rs2035580980 | 1292 | L>P | No |
TOPMed gnomAD |
|
| rs1335061632 | 1293 | R>Q | No |
TOPMed gnomAD |
|
|
COSM5462428 COSM5462426 rs367837232 |
1293 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1310793939 | 1295 | T>I | No | gnomAD | |
| rs2035582349 | 1296 | D>A | No | Ensembl | |
| rs778478182 | 1296 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1289159562 | 1297 | A>T | No | gnomAD | |
| rs757799461 | 1298 | M>I | No |
ExAC gnomAD |
|
| rs1210847045 | 1298 | M>T | No | gnomAD | |
| rs201354127 | 1298 | M>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780853666 | 1299 | K>T | No |
ExAC gnomAD |
|
|
COSM5717826 COSM5717828 |
1302 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs966747263 | 1303 | E>K | No | TOPMed | |
| rs2035583922 | 1304 | F>L | No | Ensembl | |
|
rs745327307 RCV001924928 |
1305 | R>G | No |
ClinVar ExAC dbSNP gnomAD |
|
| TCGA novel | 1305 | R>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2035584057 | 1305 | R>Q | No |
TOPMed gnomAD |
|
|
COSM3499744 COSM3499742 rs745327307 |
1305 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
COSM4053496 COSM4053498 |
1307 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1387346638 | 1307 | E>D | No | gnomAD | |
| rs575599729 | 1308 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs1163597995 | 1308 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1389600474 | 1309 | S>G | No | gnomAD | |
| rs1333289419 | 1310 | M>T | No |
TOPMed gnomAD |
|
| rs768518027 | 1312 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs774091908 | 1313 | A>T | No |
ExAC TOPMed gnomAD |
|
|
RCV002015664 rs761740855 COSM5362907 COSM5362909 |
1313 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs1032474692 | 1314 | L>M | No |
TOPMed gnomAD |
|
| rs1251097645 | 1315 | Q>* | No | gnomAD | |
| rs1337938231 | 1316 | H>Q | No | gnomAD | |
|
COSM3499747 COSM3499745 |
1317 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2035586523 | 1317 | P>T | No |
TOPMed gnomAD |
|
| rs564762598 | 1320 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs564762598 | 1320 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1343146624 | 1321 | A>T | No | TOPMed | |
|
RCV001867793 COSM124206 rs759654951 |
1321 | A>V | upper_aerodigestive_tract [Cosmic] | No |
cosmic curated ClinVar ExAC TOPMed dbSNP gnomAD |
| rs1381857016 | 1322 | L>F | No |
TOPMed gnomAD |
|
| rs764587553 | 1324 | G>A | No |
ExAC gnomAD |
|
| rs1372221915 | 1324 | G>R | No |
TOPMed gnomAD |
|
| rs1372221915 | 1324 | G>S | No |
TOPMed gnomAD |
|
| rs1172705479 | 1325 | I>T | No | gnomAD | |
| rs2035588745 | 1325 | I>V | No | Ensembl | |
|
COSM3815678 COSM3815680 |
1326 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751906540 | 1326 | S>G | No |
ExAC gnomAD |
|
| rs757854467 | 1326 | S>T | No | ExAC | |
| rs1596330624 | 1328 | H>P | No | Ensembl | |
| rs965565930 | 1328 | H>Y | No |
TOPMed gnomAD |
|
| rs1380724814 | 1329 | P>L | No | gnomAD | |
| rs1294484281 | 1330 | L>R | No | gnomAD | |
| rs1325029974 | 1331 | C>Y | No | gnomAD | |
| rs1029846060 | 1333 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs768226866 | 1335 | E>D | No |
ExAC gnomAD |
|
| rs1370626875 | 1335 | E>G | No |
TOPMed gnomAD |
|
| rs918860460 | 1336 | L>F | No | Ensembl | |
| rs918860460 | 1336 | L>V | No | Ensembl | |
| rs1483658995 | 1337 | A>T | No |
TOPMed gnomAD |
|
| rs772060404 | 1337 | A>V | No |
ExAC gnomAD |
|
| rs200335851 | 1338 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200335851 | 1338 | P>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1424097791 | 1340 | S>C | No | gnomAD | |
| rs1230648334 | 1343 | N>K | No | gnomAD | |
| rs1360380885 | 1344 | T>A | No |
TOPMed gnomAD |
|
| rs1360380885 | 1344 | T>S | No |
TOPMed gnomAD |
|
| rs2035594377 | 1345 | V>L | No | Ensembl | |
| rs1251141495 | 1347 | S>F | No | TOPMed | |
|
RCV001953961 rs779620336 |
1348 | E>A | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs750960615 | 1348 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs750960615 | 1348 | E>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1349 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2035596234 | 1350 | A>D | No | Ensembl | |
|
RCV000967395 rs77202443 |
1350 | A>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1304127213 | 1351 | R>G | No | gnomAD | |
| rs566579135 | 1352 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1567272318 | 1352 | D>N | No | Ensembl | |
| rs55727462 | 1352 | D>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1353 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1297029520 | 1356 | I>V | No | gnomAD | |
| rs1379186022 | 1357 | P>T | No | gnomAD | |
| rs1206866127 | 1360 | H>N | No | TOPMed | |
| rs1314503355 | 1366 | I>M | No | gnomAD | |
| rs1567274364 | 1366 | I>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1281760625 | 1366 | I>T | No | gnomAD | |
| rs1447669595 | 1366 | I>V | No |
TOPMed gnomAD |
|
| rs2035711710 | 1371 | A>D | No | gnomAD | |
| rs760940198 | 1371 | A>P | No |
ExAC gnomAD |
|
| rs760940198 | 1371 | A>T | No |
ExAC gnomAD |
|
| rs766947451 | 1372 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs766947451 | 1372 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs1354255288 | 1374 | L>P | No | TOPMed | |
| rs1203719103 | 1375 | A>V | No | gnomAD | |
| rs368406955 | 1376 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1178672341 | 1378 | H>Q | No | gnomAD | |
| rs758064403 | 1379 | K>N | No |
ExAC gnomAD |
|
| rs1304499816 | 1380 | K>R | No | Ensembl | |
| rs763640946 | 1381 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1183240932 | 1384 | F>I | No | gnomAD | |
|
COSM3420179 COSM3420181 |
1389 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs956247305 | 1392 | I>V | No |
TOPMed gnomAD |
|
| TCGA novel | 1393 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2035716118 | 1394 | V>L | No |
TOPMed gnomAD |
|
| rs2035716597 | 1395 | W>C | No |
TOPMed gnomAD |
|
| rs1403131690 | 1395 | W>R | No | gnomAD | |
| rs2035716747 | 1396 | S>F | No | TOPMed | |
| rs2035717159 | 1397 | L>F | No | TOPMed | |
| rs1415733977 | 1398 | D>V | No | TOPMed | |
| rs2141140546 | 1399 | V>G | No | Ensembl | |
|
RCV002009237 rs756048864 |
1399 | V>I | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs779146929 | 1400 | K>* | No |
ExAC TOPMed gnomAD |
|
| rs779146929 | 1400 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs748220756 | 1400 | K>T | No |
ExAC gnomAD |
|
| rs772400768 | 1401 | E>Q | No |
ExAC gnomAD |
|
| rs1232673293 | 1402 | H>P | No | TOPMed | |
| rs1379599094 | 1402 | H>Q | No | gnomAD | |
| rs2035719020 | 1402 | H>Y | No | TOPMed | |
| rs1213816191 | 1403 | I>V | No |
TOPMed gnomAD |
|
| rs757356517 | 1407 | L>P | No |
ExAC gnomAD |
|
| rs757356517 | 1407 | L>R | No |
ExAC gnomAD |
|
| rs1261894228 | 1409 | D>E | No | gnomAD | |
| rs2035721261 | 1409 | D>G | No | gnomAD | |
| rs41527944 | 1410 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1211781027 RCV001890815 |
1411 | G>R | No |
ClinVar dbSNP gnomAD |
|
| rs1254240956 | 1412 | I>F | No | TOPMed | |
|
COSM3499753 rs558517232 COSM3499751 |
1413 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA gnomAD |
|
COSM215599 rs763849093 |
1421 | A>T | central_nervous_system [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM2156666 COSM2156664 rs767219901 |
1424 | V>M | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs750184775 | 1425 | E>G | No |
ExAC gnomAD |
|
| rs1296053613 | 1425 | E>K | No | gnomAD | |
| rs1038771788 | 1428 | P>T | No | TOPMed | |
| rs780131853 | 1429 | G>A | No |
ExAC gnomAD |
|
| rs780131853 | 1429 | G>D | No |
ExAC gnomAD |
|
| TCGA novel | 1430 | Y>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1326547383 | 1434 | E>* | No | Ensembl | |
| rs752860998 | 1434 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs1348226308 | 1437 | P>S | No |
TOPMed gnomAD |
|
|
rs199787324 RCV001981432 |
1438 | R>C | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs777857679 | 1438 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs371409478 | 1439 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1157031419 | 1440 | V>I | No |
TOPMed gnomAD |
|
| rs2035728059 | 1443 | E>Q | No | Ensembl | |
| rs771887826 | 1446 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs772974207 | 1447 | M>I | No |
ExAC gnomAD |
|
| rs1443987546 | 1450 | Y>C | No |
TOPMed gnomAD |
|
| rs1448841245 | 1452 | M>I | No |
TOPMed gnomAD |
|
| rs2035840467 | 1452 | M>T | No | gnomAD | |
| rs61733306 | 1455 | Y>* | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1408511714 | 1456 | E>K | No | gnomAD | |
|
TCGA novel rs2035842834 |
1462 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1330683300 | 1462 | R>H | No |
TOPMed gnomAD |
|
| rs765212080 | 1463 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs751661175 | 1464 | A>V | No |
ExAC gnomAD |
|
| rs559486146 | 1468 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs756290924 | 1471 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs750667752 | 1471 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs780409265 | 1472 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs2035844672 | 1474 | K>E | No | TOPMed | |
| rs1567276891 | 1475 | K>N | No |
TOPMed gnomAD |
|
| rs1399409229 | 1478 | K>N | No | gnomAD | |
| rs755499854 | 1478 | K>R | No |
ExAC TOPMed gnomAD |
|
|
COSM699621 COSM1646646 |
1480 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377318134 | 1480 | I>N | No |
ESP ExAC TOPMed gnomAD |
|
| COSM959638 | 1481 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747660022 | 1481 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001965690 rs2035845790 |
1482 | P>L | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs2035845790 | 1482 | P>Q | No |
TOPMed gnomAD |
|
| rs2035846282 | 1485 | G>R | No | TOPMed | |
| rs201964335 | 1487 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1197526686 | 1488 | E>K | No | gnomAD | |
| rs1197526686 | 1488 | E>Q | No | gnomAD | |
| rs1246033369 | 1489 | E>* | No | gnomAD | |
| rs1277995966 | 1489 | E>D | No | gnomAD | |
| rs1201085096 | 1491 | Q>K | No |
1000Genomes gnomAD |
|
| rs367806940 | 1493 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
|
rs367806940 RCV001906415 |
1493 | R>Q | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs374925554 | 1493 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs775480216 | 1494 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs775480216 | 1494 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs775480216 | 1494 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1187939898 | 1495 | L>M | No | gnomAD | |
| rs2141144473 | 1496 | Q>R | No | Ensembl | |
|
rs767617347 COSM171440 |
1497 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs756382869 | 1498 | L>F | No |
ExAC gnomAD |
|
| rs376349366 | 1500 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs755347885 | 1501 | E>K | No |
ExAC gnomAD |
|
| rs1396270675 | 1502 | C>* | No |
TOPMed gnomAD |
|
| rs2035849580 | 1503 | W>* | No | TOPMed | |
| rs779096886 | 1505 | T>S | No |
ExAC gnomAD |
|
| rs1220993308 | 1507 | P>S | No | gnomAD | |
| rs1378059560 | 1510 | R>* | No |
TOPMed gnomAD |
|
| rs1378059560 | 1510 | R>G | No |
TOPMed gnomAD |
|
| rs2035912573 | 1510 | R>Q | No | TOPMed | |
| rs909700518 | 1511 | P>L | No |
TOPMed gnomAD |
|
| rs749075771 | 1512 | L>P | No |
ExAC gnomAD |
|
| rs749075771 | 1512 | L>Q | No |
ExAC gnomAD |
|
| rs1349671116 | 1513 | A>D | No | gnomAD | |
| rs761689956 | 1516 | V>A | No |
ExAC gnomAD |
|
| rs2035913766 | 1516 | V>M | No | TOPMed | |
| rs771073835 | 1517 | V>M | No |
ExAC gnomAD |
|
| rs765366973 | 1518 | S>R | No |
ExAC gnomAD |
|
| rs759816169 | 1518 | S>T | No | ExAC | |
| rs2035914868 | 1520 | M>I | No | Ensembl | |
| rs753181668 | 1521 | K>M | No |
ExAC TOPMed gnomAD |
|
|
RCV001929237 rs753181668 |
1521 | K>R | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs2035915220 | 1523 | P>L | No |
TOPMed gnomAD |
|
|
COSM4932196 COSM4932194 |
1525 | F>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764746962 | 1526 | A>V | No |
ExAC gnomAD |
|
| TCGA novel | 1527 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs369644113 | 1527 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV002010190 rs2141146629 |
1528 | F>L | No |
ClinVar Ensembl dbSNP |
|
| rs1436564385 | 1529 | M>T | No | gnomAD | |
|
COSM2200724 COSM959640 |
1529 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2035916292 | 1530 | Y>C | No | Ensembl | |
| rs2141146653 | 1533 | C>R | No | Ensembl | |
| TCGA novel | 1534 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2141146664 | 1534 | C>Y | No | 1000Genomes | |
| rs1347163852 | 1536 | K>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1537 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1177495292 | 1538 | T>A | No |
TOPMed gnomAD |
|
| rs2035917156 | 1539 | A>V | No | TOPMed | |
| rs377128940 | 1540 | F>L | No |
ESP TOPMed gnomAD |
|
| rs749127854 | 1540 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs779646604 | 1540 | F>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4827086 COSM4827084 |
1542 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM282561 | 1543 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1345169860 | 1543 | S>P | No |
TOPMed gnomAD |
|
| rs1373624280 | 1545 | G>C | No | Ensembl | |
| rs1373624280 | 1545 | G>R | No | Ensembl | |
| rs1465581413 | 1546 | Q>R | No | gnomAD | |
| rs1316812128 | 1547 | E>D | No |
TOPMed gnomAD |
|
| rs754789492 | 1548 | Y>N | No |
ExAC gnomAD |
|
| rs778638365 | 1549 | T>A | No |
ExAC gnomAD |
|
| rs1378137531 | 1550 | V>M | No |
TOPMed gnomAD |
|
| rs2035919622 | 1552 | F>L | No | TOPMed | |
| rs771844282 | 1554 | D>N | No |
ExAC gnomAD |
|
| rs776831970 | 1556 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1567278927 | 1556 | K>N | No | Ensembl | |
| rs1335332740 | 1558 | E>D | No | gnomAD | |
|
COSM699619 COSM1646645 |
1558 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs531262021 | 1559 | S>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778249403 | 1559 | S>Y | No | gnomAD | |
|
COSM5178767 COSM1375696 |
1560 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774839951 | 1563 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1357322729 | 1563 | T>P | No |
TOPMed gnomAD |
|
| rs2036154581 | 1564 | V>E | No | TOPMed | |
| rs1364955563 | 1566 | N>D | No | gnomAD | |
| rs2036155502 | 1568 | E>K | No |
TOPMed gnomAD |
|
| rs2036155635 | 1569 | K>T | No | Ensembl | |
| rs1220581195 | 1570 | G>R | No | gnomAD | |
| rs761200124 | 1571 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1288624627 | 1572 | M>I | No | gnomAD | |
|
COSM6076713 COSM6076715 |
1572 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1214844164 | 1572 | M>R | No |
TOPMed gnomAD |
|
| rs765878640 | 1572 | M>V | No |
ExAC gnomAD |
|
| rs371591965 | 1574 | V>L | No |
ESP TOPMed gnomAD |
|
| rs371591965 | 1574 | V>M | No |
ESP TOPMed gnomAD |
|
| rs61733307 | 1577 | M>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1577 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1484972951 | 1578 | C>Y | No | TOPMed | |
| rs2036157220 | 1579 | C>S | No | Ensembl | |
| rs1194473412 | 1580 | P>S | No | gnomAD | |
| COSM1300963 | 1582 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1379874070 | 1582 | M>L | No |
TOPMed gnomAD |
|
| rs1156549086 | 1583 | K>N | No | gnomAD | |
| rs752579784 | 1583 | K>R | No |
ExAC gnomAD |
|
|
RCV001955771 rs2141154262 |
1585 | S>N | No |
ClinVar Ensembl dbSNP |
|
| rs758106459 | 1585 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs757198351 | 1587 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1442511934 | 1589 | Q>* | No | gnomAD | |
| rs2141154307 | 1589 | Q>R | No | Ensembl | |
| rs749368396 | 1590 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs2036159011 | 1590 | V>I | No |
TOPMed gnomAD |
|
| rs2036159501 | 1591 | Q>R | No | TOPMed | |
| rs769036375 | 1593 | S>C | No |
ExAC gnomAD |
|
| rs1311911108 | 1593 | S>P | No | gnomAD | |
| rs2036159888 | 1594 | L>M | No | TOPMed | |
| rs2141154365 | 1594 | L>R | No | Ensembl | |
| rs748349627 | 1597 | A>G | No |
ExAC gnomAD |
|
| rs748349627 | 1597 | A>V | No |
ExAC gnomAD |
|
| rs1214343969 | 1598 | T>I | No | gnomAD | |
| rs773558472 | 1599 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs747368593 | 1600 | D>H | No |
ExAC gnomAD |
|
| rs747368593 | 1600 | D>N | No |
ExAC gnomAD |
|
| rs777102768 | 1601 | Q>E | No |
ExAC gnomAD |
|
|
RCV001910683 rs759987456 |
1601 | Q>L | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs759987456 | 1601 | Q>R | No |
ExAC gnomAD |
|
| rs2141156873 | 1604 | Y>H | No | Ensembl | |
| rs779968095 | 1605 | I>M | No | Ensembl | |
| rs1253802771 | 1607 | T>I | No |
TOPMed gnomAD |
|
| TCGA novel | 1608 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2036250396 | 1608 | L>V | No | TOPMed | |
| rs1255284630 | 1612 | C>S | No |
TOPMed gnomAD |
|
| rs761675199 | 1615 | N>S | No |
ExAC gnomAD |
|
| rs2141156925 | 1616 | T>A | No | Ensembl | |
| rs1465226737 | 1616 | T>I | No | gnomAD | |
| rs1465226737 | 1616 | T>K | No | gnomAD | |
| rs767370051 | 1617 | P>S | No |
ExAC gnomAD |
|
| rs2036251893 | 1618 | Q>K | No |
TOPMed gnomAD |
|
| rs756045028 | 1619 | Q>H | No |
ExAC gnomAD |
|
| rs2036252196 | 1619 | Q>K | No | Ensembl | |
| rs2141156955 | 1619 | Q>R | No | Ensembl | |
| rs765244575 | 1620 | A>V | No |
ExAC gnomAD |
|
| rs1567285251 | 1621 | L>W | No | Ensembl | |
| TCGA novel | 1622 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1000097370 | 1624 | P>L | No | Ensembl | |
| rs903026949 | 1624 | P>S | No |
TOPMed gnomAD |
|
| rs753877975 | 1626 | V>A | No | gnomAD | |
|
COSM4970009 COSM4970011 rs778024046 |
1627 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1033347216 | 1629 | C>S | No | Ensembl | |
| rs2036254147 | 1631 | L>* | No | gnomAD | |
|
COSM5178768 COSM1375698 |
1632 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs552313052 | 1633 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1484812789 | 1634 | P>A | No | gnomAD | |
| rs1567285333 | 1634 | P>L | No | Ensembl | |
| TCGA novel | 1634 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2036255295 | 1635 | V>A | No | Ensembl | |
| rs932536919 | 1636 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
RCV001355138 rs746379912 |
1637 | K>* | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs746379912 | 1637 | K>E | No |
ExAC gnomAD |
|
| COSM959642 | 1637 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770281170 | 1637 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs2036468764 | 1640 | S>P | No |
TOPMed gnomAD |
|
| rs1471018022 | 1642 | L>R | No |
TOPMed gnomAD |
|
| rs2036469044 | 1643 | V>I | No | Ensembl | |
| rs1359787858 | 1645 | A>V | No |
TOPMed gnomAD |
|
| rs1412941091 | 1646 | G>D | No |
TOPMed gnomAD |
|
| rs572219729 | 1648 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs572219729 RCV002014704 |
1648 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar 1000Genomes ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs764163827 | 1649 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1185932993 | 1651 | L>P | No | Ensembl | |
| rs762084426 | 1652 | V>L | No |
ExAC gnomAD |
|
| rs767727219 | 1653 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs750797544 | 1653 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs76666049 | 1654 | V>G | No |
ExAC gnomAD |
|
| rs754370308 | 1655 | F>V | No |
ExAC gnomAD |
|
| rs74775581 | 1656 | P>A | No |
ExAC gnomAD |
|
| rs74775581 | 1656 | P>S | No |
ExAC gnomAD |
|
| TCGA novel | 1657 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200281447 | 1657 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs370180202 | 1659 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs370180202 | 1659 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4053507 COSM4053505 rs1015630432 |
1659 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs770791811 | 1660 | G>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1660 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs770791811 RCV001998392 |
1660 | G>D | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1412835234 | 1660 | G>S | No | gnomAD | |
| rs1596359228 | 1661 | T>A | No | Ensembl | |
| rs769709976 | 1661 | T>N | No |
ExAC gnomAD |
|
| rs1596359228 | 1661 | T>P | No | Ensembl | |
| rs373755921 | 1662 | P>L | No |
ESP ExAC gnomAD |
|
| rs570984561 | 1664 | D>N | No | Ensembl | |
| rs1484921427 | 1665 | S>C | No | gnomAD | |
| rs1484921427 | 1665 | S>G | No | gnomAD | |
| rs1190652512 | 1665 | S>R | No |
TOPMed gnomAD |
|
| rs750686458 | 1666 | C>Y | No |
ExAC gnomAD |
|
| rs760737980 | 1667 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs760737980 | 1667 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2036475933 | 1668 | Y>* | No | Ensembl | |
| rs2036475790 | 1668 | Y>S | No | gnomAD | |
|
rs2141163286 RCV001929049 |
1670 | C>G | No |
ClinVar Ensembl dbSNP |
|
| rs2141163286 | 1670 | C>S | No | Ensembl | |
|
COSM1375700 COSM5130294 |
1670 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1302269451 | 1671 | S>* | No |
TOPMed gnomAD |
|
| rs371053735 | 1671 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs555149433 | 1672 | H>Q | No |
TOPMed gnomAD |
|
| rs375652666 | 1672 | H>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs779529883 | 1674 | A>T | No |
ExAC gnomAD |
|
| rs2036476787 | 1676 | R>S | No | TOPMed | |
| rs1356555427 | 1677 | S>F | No | TOPMed | |
| rs2036477041 | 1678 | K>R | No | TOPMed | |
| TCGA novel | 1679 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2036477334 | 1680 | S>N | No | Ensembl | |
|
COSM959646 rs777107638 |
1682 | A>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
RCV001904355 rs368322542 |
1682 | A>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs745649117 | 1683 | D>Y | No |
ExAC gnomAD |
|
| rs1166044505 | 1685 | D>G | No | TOPMed | |
| rs370749356 | 1686 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370749356 | 1686 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772139995 | 1687 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs373893782 | 1687 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1213403128 | 1689 | N>I | No |
TOPMed gnomAD |
|
| rs1281510100 | 1690 | P>L | No | gnomAD | |
| rs2036479949 | 1691 | Y>C | No | Ensembl | |
| rs2036479708 | 1691 | Y>H | No | Ensembl | |
| rs2036480477 | 1694 | K>Q | No | Ensembl | |
| rs2036480943 | 1696 | M>I | No | Ensembl | |
| rs957642850 | 1696 | M>T | No | TOPMed | |
| rs189062191 | 1700 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs189062191 | 1700 | N>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs913687680 | 1702 | G>D | No | Ensembl | |
|
COSM1735675 RCV002088628 rs78605716 |
1702 | G>S | pancreas [Cosmic] | No |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs372292557 RCV002026000 |
1703 | S>Y | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1400025585 | 1705 | V>I | No | gnomAD | |
| rs1596359545 | 1710 | G>E | No | Ensembl | |
| rs758909087 | 1711 | P>L | No |
ExAC gnomAD |
|
| rs1316190212 | 1711 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs751022115 | 1715 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2036483848 | 1717 | D>A | No |
TOPMed gnomAD |
|
| rs780680269 | 1717 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs745716119 | 1718 | C>F | No | ExAC | |
| rs1304619504 | 1719 | A>T | No | gnomAD | |
| rs2036484338 | 1719 | A>V | No | Ensembl | |
| rs76740676 | 1720 | S>P | No | Ensembl | |
| rs1222535802 | 1724 | C>S | No | gnomAD | |
| rs2036485106 | 1725 | R>G | No | TOPMed | |
| rs779753081 | 1726 | R>L | No |
ExAC gnomAD |
|
| rs779753081 | 1726 | R>Q | No |
ExAC gnomAD |
|
| rs77374252 | 1726 | R>W | No | Ensembl | |
| rs2141163568 | 1727 | L>V | No | Ensembl | |
| rs2141163581 | 1728 | E>G | No | Ensembl | |
| rs1194569505 | 1730 | Y>* | No |
TOPMed gnomAD |
|
| rs374196433 | 1730 | Y>C | No |
ESP ExAC gnomAD |
|
| rs2141163590 | 1730 | Y>H | No | Ensembl | |
| rs771231472 | 1731 | M>T | No |
ExAC gnomAD |
|
|
COSM4932755 COSM4932757 rs921178428 |
1731 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1469472150 | 1732 | A>V | No | gnomAD | |
| rs2141163629 | 1733 | P>S | No | Ensembl | |
| rs775143193 | 1735 | M>I | No |
TOPMed gnomAD |
|
| rs765702635 | 1735 | M>T | No |
ExAC TOPMed gnomAD |
|
|
rs3829706 RCV000879392 |
1735 | M>V | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1391730839 | 1737 | T>A | No | gnomAD | |
|
COSM1640190 rs939725500 COSM1640192 |
1737 | T>M | Variant assessed as Somatic; MODERATE impact. stomach [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs2036487593 | 1738 | S>* | No | gnomAD | |
|
COSM959650 rs763615713 |
1740 | V>M | endometrium [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs549997085 | 1741 | C>* | No |
1000Genomes ExAC gnomAD |
|
| rs1228022677 | 1742 | S>N | No | TOPMed | |
| rs1351947491 | 1742 | S>R | No | gnomAD | |
| rs1348798423 | 1744 | E>G | No |
TOPMed gnomAD |
|
| rs751124500 | 1744 | E>K | No |
ExAC gnomAD |
|
| rs756702653 | 1745 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs1337102657 | 1745 | G>D | No | gnomAD | |
| rs756702653 | 1745 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs2036489109 | 1747 | G>R | No | Ensembl | |
| rs1390431435 | 1748 | E>K | No | TOPMed | |
| rs750110863 | 1748 | E>V | No | ExAC | |
| rs755701846 | 1750 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs755701846 | 1750 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs754980135 | 1751 | V>G | No |
ExAC gnomAD |
|
| rs373452117 | 1751 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1036301171 | 1752 | W>L | No | TOPMed | |
| rs1409394167 | 1754 | L>M | No |
TOPMed gnomAD |
|
| rs1023161317 | 1756 | D>N | No | Ensembl | |
| rs1165040188 | 1757 | K>Q | No | gnomAD | |
| rs766954384 | 1759 | N>S | No | TOPMed | |
| rs766954384 | 1759 | N>T | No | TOPMed | |
| rs2141163805 | 1760 | S>A | No | Ensembl | |
| rs770946839 | 1760 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs1331291177 | 1761 | L>F | No | gnomAD | |
| rs373287839 | 1763 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs770129962 | 1763 | M>T | No |
ExAC gnomAD |
|
| rs1003141890 | 1766 | S>F | No | Ensembl | |
| rs1357947247 | 1767 | T>I | No |
TOPMed gnomAD |
|
| rs763381102 | 1768 | T>A | No |
ExAC gnomAD |
|
| rs763381102 | 1768 | T>P | No |
ExAC gnomAD |
|
| rs2141163866 | 1768 | T>S | No | Ensembl | |
| TCGA novel | 1770 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762344317 | 1770 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2036493750 | 1771 | L>P | No | gnomAD | |
| rs2036494044 | 1773 | A>G | No |
TOPMed gnomAD |
|
| rs2036494044 | 1773 | A>V | No |
TOPMed gnomAD |
|
| rs201726240 | 1774 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749917918 | 1777 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs146084570 | 1778 | G>R | No |
1000Genomes ExAC gnomAD |
|
|
rs3764738 RCV002210770 |
1779 | V>D | No |
ClinVar 1000Genomes ExAC dbSNP gnomAD |
|
| rs751445121 | 1779 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs779167127 | 1780 | P>S | No |
ExAC gnomAD |
|
| rs1299597590 | 1781 | S>T | No | Ensembl | |
| rs2141163958 | 1784 | R>G | No | Ensembl | |
| rs1397616623 | 1784 | R>T | No | gnomAD | |
|
COSM5135565 COSM1375702 |
1786 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2036496378 | 1786 | M>T | No |
TOPMed gnomAD |
|
| TCGA novel | 1786 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs745940472 | 1789 | V>A | No |
ExAC gnomAD |
|
| rs536296948 | 1789 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM470423 rs536296948 |
1789 | V>M | kidney Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs749481480 | 1790 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs780403978 | 1790 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs768937350 | 1791 | P>S | No |
ExAC gnomAD |
|
| rs2036497816 | 1793 | D>V | No | gnomAD | |
| rs1270951065 | 1793 | D>Y | No |
TOPMed gnomAD |
|
| rs368408781 | 1794 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001997559 rs772559934 |
1794 | T>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs368408781 | 1794 | T>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1596360339 | 1795 | E>D | No | Ensembl | |
|
rs753519057 RCV002016988 |
1797 | P>L | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1333712810 | 1797 | P>S | No | gnomAD | |
| TCGA novel | 1798 | A>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764953988 | 1798 | A>T | No |
ExAC gnomAD |
|
| rs752524474 | 1802 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs752524474 | 1802 | T>R | No |
ExAC TOPMed gnomAD |
|
| VAR_040674 | 1803 | A>T | No | UniProt | |
| rs912178976 | 1804 | N>D | No | gnomAD | |
| rs200557780 | 1804 | N>K | No |
TOPMed gnomAD |
|
| rs750583241 | 1804 | N>S | No |
ExAC gnomAD |
|
| rs199974382 | 1805 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1485930820 | 1807 | V>A | No | gnomAD | |
|
COSM4833774 COSM4833776 rs749480379 |
1809 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2036501336 | 1810 | G>E | No | gnomAD | |
| rs2036501221 | 1810 | G>R | No | gnomAD | |
| rs1319081084 | 1811 | D>Y | No |
TOPMed gnomAD |
|
| rs768906258 | 1812 | S>P | No |
ExAC TOPMed gnomAD |
|
|
rs35787282 RCV002135663 |
1814 | A>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1349850721 | 1814 | A>V | No | gnomAD | |
| rs2036502391 | 1815 | D>Y | No | TOPMed | |
|
rs377185660 RCV001940229 |
1816 | V>M | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2036502966 | 1817 | S>T | No | Ensembl | |
| rs1248109013 | 1818 | I>V | No |
TOPMed gnomAD |
|
| rs1455010578 | 1819 | M>I | No | gnomAD | |
| rs764939883 | 1819 | M>T | No |
ExAC TOPMed gnomAD |
|
|
COSM959652 rs368591954 |
1819 | M>V | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2141164235 | 1820 | Y>* | No | Ensembl | |
| rs1158867051 | 1821 | S>G | No |
TOPMed gnomAD |
|
| VAR_040675 | 1824 | L>F | No | UniProt | |
| rs2141164255 | 1824 | L>R | No | Ensembl | |
| rs775190189 | 1826 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1372045084 | 1830 | I>M | No | gnomAD | |
| rs542657977 | 1830 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1441885748 | 1831 | H>Q | No |
TOPMed gnomAD |
|
| rs1387083676 | 1831 | H>Y | No |
TOPMed gnomAD |
|
| rs757198165 | 1832 | Q>E | No |
ExAC TOPMed gnomAD |
|
|
RCV000953700 rs35128996 |
1835 | L>F | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs755191615 | 1837 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs755191615 | 1837 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1483594310 | 1839 | C>G | No | TOPMed | |
| rs779170016 | 1841 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2036506840 | 1841 | M>V | No | Ensembl | |
| rs1254447279 | 1842 | S>Y | No |
TOPMed gnomAD |
|
| rs1359017884 | 1844 | Y>N | No | TOPMed | |
| rs1285277039 | 1845 | S>Y | No | TOPMed | |
| rs201730508 | 1846 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| VAR_040676 | 1847 | S>N | No | UniProt | |
| rs968761875 | 1849 | P>R | No | TOPMed | |
| rs1021604666 | 1849 | P>S | No | gnomAD | |
|
RCV000893714 rs41525944 |
1850 | R>C | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM554333 rs200771530 RCV002143539 |
1850 | R>H | lung [Cosmic] | No |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs200771530 | 1850 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs41525944 | 1850 | R>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1167224033 | 1852 | A>P | No |
TOPMed gnomAD |
|
| rs776071685 | 1853 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs776071685 | 1853 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs769383028 | 1854 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs2036509650 | 1854 | R>T | No | TOPMed | |
| rs61733308 | 1855 | S>A | No |
TOPMed gnomAD |
|
|
RCV001997772 rs2141164457 |
1855 | S>C | No |
ClinVar Ensembl dbSNP |
|
| rs61733308 | 1855 | S>P | No |
TOPMed gnomAD |
|
| rs61733308 | 1855 | S>T | No |
TOPMed gnomAD |
|
| rs1393040389 | 1856 | P>L | No | gnomAD | |
| rs1393040389 | 1856 | P>R | No | gnomAD | |
| rs1415696374 | 1857 | S>T | No | TOPMed | |
|
rs34874770 RCV002191681 |
1858 | S>N | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1251572799 | 1860 | P>S | No |
TOPMed gnomAD |
|
| rs774234138 | 1861 | S>G | No | ExAC | |
| rs761904263 | 1861 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs767454605 | 1863 | P>S | No |
ExAC gnomAD |
|
| rs753910283 | 1864 | A>P | No |
ExAC gnomAD |
|
| rs1381987505 | 1865 | S>G | No | gnomAD | |
| rs1396906691 | 1868 | S>T | No | gnomAD | |
| TCGA novel | 1869 | V>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2036512516 | 1869 | V>M | No | TOPMed | |
| rs758557491 | 1870 | P>L | No | ExAC | |
| rs752966389 | 1870 | P>S | No |
ExAC gnomAD |
|
| TCGA novel | 1871 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778168227 | 1873 | T>S | No |
ExAC gnomAD |
|
| rs565686156 | 1874 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1414704844 | 1875 | C>F | No |
TOPMed gnomAD |
|
| rs1414704844 | 1875 | C>Y | No |
TOPMed gnomAD |
|
| rs769380465 | 1876 | E>G | No |
ExAC TOPMed gnomAD |
|
|
rs369774424 RCV001974779 |
1876 | E>K | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs931027800 | 1879 | D>E | No |
TOPMed gnomAD |
|
| rs1463294287 | 1880 | M>K | No | gnomAD | |
| rs373539914 | 1881 | L>R | No |
ESP TOPMed |
|
| rs2036515093 | 1883 | T>A | No | Ensembl | |
| rs3764740 | 1883 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs3764740 | 1883 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1214470436 | 1884 | P>S | No | gnomAD | |
| rs2141164642 | 1885 | G>D | No | Ensembl | |
| rs377202593 | 1885 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1175694807 | 1886 | A>V | No | TOPMed | |
| COSM470425 | 1888 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1292951232 | 1889 | D>E | No | gnomAD | |
| rs770606570 | 1889 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs760461091 | 1893 | H>N | No |
ExAC TOPMed gnomAD |
|
| rs1206245367 | 1893 | H>R | No | gnomAD | |
| rs760461091 | 1893 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1484972583 | 1896 | T>I | No | gnomAD | |
| rs763228498 | 1897 | P>L | No |
ExAC gnomAD |
|
| rs370127139 | 1897 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs370127139 | 1897 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1898 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764270132 | 1898 | M>T | No |
ExAC gnomAD |
|
| rs1475147925 | 1898 | M>V | No |
TOPMed gnomAD |
|
| rs367810590 | 1899 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
|
TCGA novel rs2036519004 |
1899 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs537362756 | 1900 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2036519586 | 1901 | E>D | No | TOPMed | |
| rs756523500 | 1901 | E>G | No |
ExAC gnomAD |
|
| rs558418183 | 1901 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs558418183 | 1901 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1432762901 | 1902 | T>I | No | gnomAD | |
|
rs779676368 RCV002048897 |
1903 | F>C | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1359707750 | 1904 | S>R | No | gnomAD | |
| rs748772285 | 1905 | Q>* | No |
ExAC gnomAD |
|
| rs778727638 | 1905 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs768468458 | 1905 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs1322423534 | 1909 | A>G | No | TOPMed | |
| rs2036520882 | 1909 | A>P | No |
TOPMed gnomAD |
|
|
COSM2200812 COSM1375704 |
1909 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372723036 | 1910 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1237101451 | 1912 | I>L | No | gnomAD | |
| rs772824076 | 1912 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs770769016 | 1914 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs745598708 | 1914 | A>V | No |
TOPMed gnomAD |
|
|
rs775588013 COSM199343 |
1915 | V>I | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs775588013 | 1915 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1252115722 | 1919 | I>L | No | gnomAD | |
| rs1252115722 | 1919 | I>V | No | gnomAD | |
| rs762998404 | 1921 | V>I | No |
ExAC gnomAD |
|
| rs2036522856 | 1922 | P>R | No | TOPMed | |
| rs961288295 | 1922 | P>S | No | Ensembl | |
| rs2036522984 | 1923 | R>K | No | TOPMed | |
|
COSM3815689 COSM3815687 |
1923 | R>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2141164860 | 1923 | R>W | No | Ensembl | |
|
COSM1678714 rs780922727 |
1924 | R>C | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM4053508 rs1472884841 COSM959658 |
1924 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1472884841 | 1924 | R>P | No | gnomAD | |
| rs774451516 | 1925 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs369056591 | 1925 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| VAR_040677 | 1927 | D>G | No | UniProt | |
| rs2036547989 | 1927 | D>H | No | gnomAD | |
| TCGA novel | 1928 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2036548207 | 1929 | I>V | No | Ensembl | |
| rs201481845 | 1930 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201481845 | 1930 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1257955496 | 1931 | I>V | No |
TOPMed gnomAD |
|
| rs200916884 | 1934 | E>K | No | gnomAD | |
| rs200916884 | 1934 | E>Q | No | gnomAD | |
| rs944767940 | 1935 | K>E | No |
TOPMed gnomAD |
|
| rs1291492431 | 1936 | D>Y | No | gnomAD | |
|
rs2924835 RCV001652123 |
1938 | G>D | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
RCV002075507 rs386787404 |
1938 | G>E | No |
ClinVar Ensembl dbSNP |
|
|
COSM4053511 COSM4053509 rs765835793 |
1939 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs752310046 | 1940 | Q>R | No |
ExAC gnomAD |
|
| rs753312283 | 1941 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs753312283 | 1941 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM5507036 rs753312283 COSM5507034 |
1941 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
RCV002140868 rs373695236 |
1941 | R>W | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| TCGA novel | 1942 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756852396 | 1943 | R>* | No |
ExAC gnomAD |
|
| rs756852396 | 1943 | R>G | No |
ExAC gnomAD |
|
|
RCV001917793 rs376808685 |
1943 | R>Q | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs375414450 RCV002041286 |
1947 | V>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs375414450 | 1947 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772079009 | 1949 | K>E | No |
ExAC gnomAD |
|
| rs1296137445 | 1951 | R>* | No | gnomAD | |
| rs777863487 | 1951 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
COSM959660 rs771248375 |
1955 | P>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1396628347 | 1956 | H>R | No | gnomAD | |
| rs773797200 | 1958 | V>M | No |
ExAC gnomAD |
|
| rs766950839 | 1960 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs766950839 | 1960 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2036666358 | 1963 | A>T | No | TOPMed | |
|
RCV001914323 rs371594206 |
1964 | V>M | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM6141643 COSM6141645 |
1965 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1227064013 | 1965 | V>M | No | gnomAD | |
| TCGA novel | 1966 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1484313302 | 1967 | K>* | No | gnomAD | |
|
RCV002030757 rs200845343 |
1968 | D>N | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs765208232 | 1972 | C>Y | No |
ExAC gnomAD |
|
| rs752623884 | 1973 | T>I | No |
ExAC gnomAD |
|
| rs1183697480 | 1974 | F>V | No | gnomAD | |
| rs1414732689 | 1975 | E>D | No | gnomAD | |
| rs916569957 | 1979 | T>A | No | TOPMed | |
| rs1160827713 | 1980 | E>D | No | Ensembl | |
| rs757416596 | 1980 | E>Q | No |
ExAC gnomAD |
|
|
COSM1608139 rs781380711 |
1982 | C>Y | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1431526955 | 1984 | A>T | No | gnomAD | |
| rs1445097736 | 1985 | V>A | No | gnomAD | |
|
rs200452795 RCV002024899 |
1985 | V>I | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs200452795 | 1985 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2036669580 | 1987 | R>K | No | Ensembl | |
| rs999534552 | 1988 | G>S | No | gnomAD | |
| rs551580410 | 1991 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs769097011 | 1991 | A>V | No |
ExAC gnomAD |
|
| rs1596367302 | 1994 | F>V | No | Ensembl | |
| rs748618009 | 1995 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs748618009 | 1995 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs200799960 | 1996 | I>V | No |
1000Genomes ExAC gnomAD |
|
| rs760287550 | 1998 | Y>H | No |
ExAC TOPMed gnomAD |
|
| rs2036671093 | 1999 | Q>H | No | TOPMed | |
| rs2036671208 | 2000 | S>F | No | Ensembl | |
|
COSM1158163 rs983155331 |
2002 | E>D | pancreas [Cosmic] | No |
cosmic curated Ensembl |
| rs3752321 | 2002 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs3752321 | 2002 | E>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1451056831 | 2005 | G>S | No | gnomAD | |
| TCGA novel | 2006 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377568599 | 2006 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs373483365 | 2006 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1452560841 | 2009 | A>D | No | gnomAD | |
| rs2036673118 | 2009 | A>T | No | Ensembl | |
| rs1452560841 | 2009 | A>V | No | gnomAD | |
|
RCV002001211 rs780306812 |
2012 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs370159409 | 2012 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs370159409 | 2012 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs748705944 | 2013 | K>Q | No |
ExAC gnomAD |
|
| rs1209685115 | 2014 | R>I | No | TOPMed | |
| rs1209685115 | 2014 | R>K | No | TOPMed | |
| rs1596367537 | 2016 | R>E | No | Ensembl | |
| rs1360508923 | 2016 | R>L | No |
TOPMed gnomAD |
1 associated diseases with Q38SD2
[MIM: 615198]: Osteosclerotic metaphyseal dysplasia (OSMD)
An autosomal recessive skeletal dysplasia characterized by osteosclerosis at multiple skeletal sites, predominantly at the metaphyses of the long bones and vertebral bodies. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal recessive skeletal dysplasia characterized by osteosclerosis at multiple skeletal sites, predominantly at the metaphyses of the long bones and vertebral bodies. . Note=The disease is caused by variants affecting the gene represented in this entry.
17 regional properties for Q38SD2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 1242 - 1525 | IPR000719 |
| repeat | Leucine-rich repeat | 303 - 324 | IPR001611-1 |
| repeat | Leucine-rich repeat | 381 - 402 | IPR001611-2 |
| repeat | Leucine-rich repeat | 549 - 570 | IPR001611-3 |
| repeat | Ankyrin repeat | 119 - 148 | IPR002110-1 |
| repeat | Ankyrin repeat | 152 - 182 | IPR002110-2 |
| repeat | Ankyrin repeat | 193 - 223 | IPR002110-3 |
| repeat | Leucine-rich repeat, typical subtype | 278 - 300 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 301 - 324 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 328 - 351 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 379 - 401 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 403 - 427 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 472 - 493 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 548 - 569 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 570 - 594 | IPR003591-8 |
| domain | Roc domain | 632 - 826 | IPR020859 |
| domain | C-terminal of Roc (COR) domain | 844 - 1036 | IPR032171 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR48055 | LEUCINE-RICH REPEAT RECEPTOR PROTEIN KINASE EMS1 |
| PANTHER Subfamily | PTHR48055:SF13 | LEUCINE-RICH REPEAT SERINE_THREONINE-PROTEIN KINASE 1 |
| PANTHER Protein Class | transmembrane signal receptor | |
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| identical protein binding | Binding to an identical protein or proteins. |
| metal ion binding | Binding to a metal ion. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| bone resorption | The process in which specialized cells known as osteoclasts degrade the organic and inorganic portions of bone, and endocytose and transport the degradation products. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of peptidyl-tyrosine phosphorylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
| osteoclast development | The process whose specific outcome is the progression of a osteoclast from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate. An osteoclast is a specialized phagocytic cell associated with the absorption and removal of the mineralized matrix of bone tissue. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of canonical Wnt signaling pathway | Any process that increases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| positive regulation of intracellular signal transduction | Any process that activates or increases the frequency, rate or extent of intracellular signal transduction. |
| positive regulation of peptidyl-tyrosine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
74 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VDU3 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Bos taurus (Bovine) | SS |
| Q4TVR5 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Bos taurus (Bovine) | PR |
| Q3SZJ2 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Bos taurus (Bovine) | PR |
| Q3MHH9 | ECM2 | Extracellular matrix protein 2 | Bos taurus (Bovine) | PR |
| P58874 | OPTC | Opticin | Bos taurus (Bovine) | PR |
| Q24K06 | LRRC10 | Leucine-rich repeat-containing protein 10 | Bos taurus (Bovine) | PR |
| Q6XUX0 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Gallus gallus (Chicken) | PR |
| P83104 | Takl1 | Putative mitogen-activated protein kinase kinase kinase 7-like | Drosophila melanogaster (Fruit fly) | PR |
| Q95UN8 | slpr | Mitogen-activated protein kinase kinase kinase | Drosophila melanogaster (Fruit fly) | EV |
| Q9V780 | Lap1 | Protein lap1 | Drosophila melanogaster (Fruit fly) | PR |
| O43353 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Homo sapiens (Human) | PR |
| Q02779 | MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | Homo sapiens (Human) | SS |
| Q8NB16 | MLKL | Mixed lineage kinase domain-like protein | Homo sapiens (Human) | EV |
| Q16584 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | Homo sapiens (Human) | EV |
| P00540 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Homo sapiens (Human) | PR |
| Q5TCX8 | MAP3K21 | Mitogen-activated protein kinase kinase kinase 21 | Homo sapiens (Human) | PR |
| Q6XUX3 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Homo sapiens (Human) | PR |
| O43318 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Homo sapiens (Human) | SS |
| Q9NYL2 | MAP3K20 | Mitogen-activated protein kinase kinase kinase 20 | Homo sapiens (Human) | PR |
| P80192 | MAP3K9 | Mitogen-activated protein kinase kinase kinase 9 | Homo sapiens (Human) | SS |
| Q96NW7 | LRRC7 | Leucine-rich repeat-containing protein 7 | Homo sapiens (Human) | PR |
| Q8IWT6 | LRRC8A | Volume-regulated anion channel subunit LRRC8A | Homo sapiens (Human) | PR |
| Q9HCJ2 | LRRC4C | Leucine-rich repeat-containing protein 4C | Homo sapiens (Human) | PR |
| Q9UFC0 | LRWD1 | Leucine-rich repeat and WD repeat-containing protein 1 | Homo sapiens (Human) | PR |
| Q96L50 | LRR1 | Leucine-rich repeat protein 1 | Homo sapiens (Human) | PR |
| Q86UN2 | RTN4RL1 | Reticulon-4 receptor-like 1 | Homo sapiens (Human) | PR |
| Q8IWK6 | ADGRA3 | Adhesion G protein-coupled receptor A3 | Homo sapiens (Human) | PR |
| Q7L1W4 | LRRC8D | Volume-regulated anion channel subunit LRRC8D | Homo sapiens (Human) | PR |
| Q96FE5 | LINGO1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Homo sapiens (Human) | PR |
| Q8TDW0 | LRRC8C | Volume-regulated anion channel subunit LRRC8C | Homo sapiens (Human) | PR |
| Q80XI6 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Mus musculus (Mouse) | PR |
| Q8VDG6 | Map3k21 | Mitogen-activated protein kinase kinase kinase 21 | Mus musculus (Mouse) | PR |
| P58801 | Ripk2 | Receptor-interacting serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q62073 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Mus musculus (Mouse) | EV |
| Q3U1V8 | Map3k9 | Mitogen-activated protein kinase kinase kinase 9 | Mus musculus (Mouse) | SS |
| Q9D2Y4 | Mlkl | Mixed lineage kinase domain-like protein | Mus musculus (Mouse) | SS |
| Q9ESL4 | Map3k20 | Mitogen-activated protein kinase kinase kinase 20 | Mus musculus (Mouse) | PR |
| P00536 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Mus musculus (Mouse) | PR |
| Q66L42 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Mus musculus (Mouse) | SS |
| A6H694 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Mus musculus (Mouse) | PR |
| Q9D9Q0 | Lrrc69 | Leucine-rich repeat-containing protein 69 | Mus musculus (Mouse) | PR |
| Q8BGI7 | Lrrc39 | Leucine-rich repeat-containing protein 39 | Mus musculus (Mouse) | PR |
| Q7TT36 | Adgra3 | Adhesion G protein-coupled receptor A3 | Mus musculus (Mouse) | PR |
| P59383 | Lrrn4 | Leucine-rich repeat neuronal protein 4 | Mus musculus (Mouse) | PR |
| Q9D1T0 | Lingo1 | Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | Mus musculus (Mouse) | PR |
| Q8R502 | Lrrc8c | Volume-regulated anion channel subunit LRRC8C | Mus musculus (Mouse) | PR |
| Q80WG5 | Lrrc8a | Volume-regulated anion channel subunit LRRC8A | Mus musculus (Mouse) | PR |
| Q8K0S5 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Mus musculus (Mouse) | PR |
| Q80TE7 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Mus musculus (Mouse) | PR |
| Q5DU41 | Lrrc8b | Volume-regulated anion channel subunit LRRC8B | Mus musculus (Mouse) | PR |
| Q5RKR3 | Islr2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | Mus musculus (Mouse) | PR |
| Q8C031 | Lrrc4c | Leucine-rich repeat-containing protein 4C | Mus musculus (Mouse) | PR |
| P0C8E4 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Rattus norvegicus (Rat) | SS |
| Q66HA1 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Rattus norvegicus (Rat) | PR |
| D3ZG83 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Rattus norvegicus (Rat) | SS |
| P00539 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Rattus norvegicus (Rat) | PR |
| P70587 | Lrrc7 | Leucine-rich repeat-containing protein 7 | Rattus norvegicus (Rat) | PR |
| Q4V8G0 | Lrrc63 | Leucine-rich repeat-containing protein 63 | Rattus norvegicus (Rat) | PR |
| Q80WD0 | Rtn4rl1 | Reticulon-4 receptor-like 1 | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9FPR3 | EDR1 | Serine/threonine-protein kinase EDR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q05609 | CTR1 | Serine/threonine-protein kinase CTR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SHI4 | RLP3 | Receptor-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5G5E0 | PIRL5 | Plant intracellular Ras-group-related LRR protein 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGE4 | At1g12460 | Probable LRR receptor-like serine/threonine-protein kinase At1g12460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0WR59 | At5g10020 | Probable inactive receptor kinase At5g10020 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q67E00 | dstyk | Dual serine/threonine and tyrosine protein kinase | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| B0JZ65 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q68F79 | lrrc8e | Volume-regulated anion channel subunit LRRC8E | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| B0R160 | lrwd1 | Leucine-rich repeat and WD repeat-containing protein 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAGMSQRPPS | MYWCVGPEES | AVCPERAMET | LNGAGDTGGK | PSTRGGDPAA | RSRRTEGIRA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AYRRGDRGGA | RDLLEEACDQ | CASQLEKGQL | LSIPAAYGDL | EMVRYLLSKR | LVELPTEPTD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DNPAVVAAYF | GHTAVVQELL | ESLPGPCSPQ | RLLNWMLALA | CQRGHLGVVK | LLVLTHGADP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ESYAVRKNEF | PVIVRLPLYA | AIKSGNEDIA | IFLLRHGAYF | CSYILLDSPD | PSKHLLRKYF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IEASPLPSSY | PGKTALRVKW | SHLRLPWVDL | DWLIDISCQI | TELDLSANCL | ATLPSVIPWG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LINLRKLNLS | DNHLGELPGV | QSSDEIICSR | LLEIDISSNK | LSHLPPGFLH | LSKLQKLTAS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KNCLEKLFEE | ENATNWIGLR | KLQELDISDN | KLTELPALFL | HSFKSLNSLN | VSRNNLKVFP |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DPWACPLKCC | KASRNALECL | PDKMAVFWKN | HLKDVDFSEN | ALKEVPLGLF | QLDALMFLRL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QGNQLAALPP | QEKWTCRQLK | TLDLSRNQLG | KNEDGLKTKR | IAFFTTRGRQ | RSGTEAASVL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| EFPAFLSESL | EVLCLNDNHL | DTVPPSVCLL | KSLSELYLGN | NPGLRELPPE | LGQLGNLWQL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| DTEDLTISNV | PAEIQKEGPK | AMLSYLRAQL | RKAEKCKLMK | MIIVGPPRQG | KSTLLEILQT |
| 670 | 680 | 690 | 700 | 710 | 720 |
| GRAPQVVHGE | ATIRTTKWEL | QRPAGSRAKV | ESVEFNVWDI | GGPASMATVN | QCFFTDKALY |
| 730 | 740 | 750 | 760 | 770 | 780 |
| VVVWNLALGE | EAVANLQFWL | LNIEAKAPNA | VVLVVGTHLD | LIEAKFRVER | IATLRAYVLA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LCRSPSGSRA | TGFPDITFKH | LHEISCKSLE | GQEGLRQLIF | HVTCSMKDVG | STIGCQRLAG |
| 850 | 860 | 870 | 880 | 890 | 900 |
| RLIPRSYLSL | QEAVLAEQQR | RSRDDDVQYL | TDRQLEQLVE | QTPDNDIKDY | EDLQSAISFL |
| 910 | 920 | 930 | 940 | 950 | 960 |
| IETGTLLHFP | DTSHGLRNLY | FLDPIWLSEC | LQRIFNIKGS | RSVAKNGVIR | AEDLRMLLVG |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| TGFTQQTEEQ | YFQFLAKFEI | ALPVANDSYL | LPHLLPSKPG | LDTHGMRHPT | ANTIQRVFKM |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| SFVPVGFWQR | FIARMLISLA | EMDLQLFENK | KNTKSRNRKV | TIYSFTGNQR | NRCSTFRVKR |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| NQTIYWQEGL | LVTFDGGYLS | VESSDVNWKK | KKSGGMKIVC | QSEVRDFSAM | AFITDHVNSL |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| IDQWFPALTA | TESDGTPLME | QYVPCPVCET | AWAQHTDPSE | KSEDVQYFDM | EDCVLTAIER |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| DFISCPRHPD | LPVPLQELVP | ELFMTDFPAR | LFLENSKLEH | SEDEGSVLGQ | GGSGTVIYRA |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| RYQGQPVAVK | RFHIKKFKNF | ANVPADTMLR | HLRATDAMKN | FSEFRQEASM | LHALQHPCIV |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| ALIGISIHPL | CFALELAPLS | SLNTVLSENA | RDSSFIPLGH | MLTQKIAYQI | ASGLAYLHKK |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| NIIFCDLKSD | NILVWSLDVK | EHINIKLSDY | GISRQSFHEG | ALGVEGTPGY | QAPEIRPRIV |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| YDEKVDMFSY | GMVLYELLSG | QRPALGHHQL | QIAKKLSKGI | RPVLGQPEEV | QFRRLQALMM |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| ECWDTKPEKR | PLALSVVSQM | KDPTFATFMY | ELCCGKQTAF | FSSQGQEYTV | VFWDGKEESR |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| NYTVVNTEKG | LMEVQRMCCP | GMKVSCQLQV | QRSLWTATED | QKIYIYTLKG | MCPLNTPQQA |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| LDTPAVVTCF | LAVPVIKKNS | YLVLAGLADG | LVAVFPVVRG | TPKDSCSYLC | SHTANRSKFS |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| IADEDARQNP | YPVKAMEVVN | SGSEVWYSNG | PGLLVIDCAS | LEICRRLEPY | MAPSMVTSVV |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| CSSEGRGEEV | VWCLDDKANS | LVMYHSTTYQ | LCARYFCGVP | SPLRDMFPVR | PLDTEPPAAS |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| HTANPKVPEG | DSIADVSIMY | SEELGTQILI | HQESLTDYCS | MSSYSSSPPR | QAARSPSSLP |
| 1870 | 1880 | 1890 | 1900 | 1910 | 1920 |
| SSPASSSSVP | FSTDCEDSDM | LHTPGAASDR | SEHDLTPMDG | ETFSQHLQAV | KILAVRDLIW |
| 1930 | 1940 | 1950 | 1960 | 1970 | 1980 |
| VPRRGGDVIV | IGLEKDSGAQ | RGRVIAVLKA | RELTPHGVLV | DAAVVAKDTV | VCTFENENTE |
| 1990 | 2000 | 2010 | |||
| WCLAVWRGWG | AREFDIFYQS | YEELGRLEAC | TRKRR |