Q2NL05
Gene name |
KIF2A |
Protein name |
Kinesin-like protein KIF2A |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:768014 |
EC number |
|
Protein Class |
|
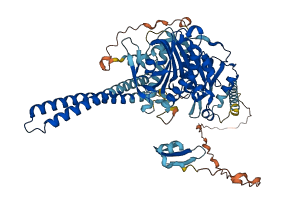
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q2NL05
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q2NL05-F1 | Predicted | AlphaFoldDB |
No variants for Q2NL05
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q2NL05 | |||||
No associated diseases with Q2NL05
Functions
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| microtubule organizing center | An intracellular structure that can catalyze gamma-tubulin-dependent microtubule nucleation and that can anchor microtubules by interacting with their minus ends, plus ends or sides. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| microtubule binding | Binding to a microtubule, a filament composed of tubulin monomers. |
| microtubule motor activity | A motor activity that generates movement along a microtubule, driven by ATP hydrolysis. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| microtubule depolymerization | The removal of tubulin heterodimers from one or both ends of a microtubule. |
| microtubule-based movement | A microtubule-based process that results in the movement of organelles, other microtubules, or other cellular components. Examples include motor-driven movement along microtubules and movement driven by polymerization or depolymerization of microtubules. |
| mitotic spindle assembly | Mitotic bipolar spindle assembly begins with spindle microtubule nucleation from the separated spindle pole body, includes spindle elongation during prometaphase, and is complete when all kinetochores are stably attached the spindle, and the spindle assembly checkpoint is satisfied. |
| mitotic spindle organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the microtubule spindle during a mitotic cell cycle. |
| nervous system development | The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state. |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVTSLNEDNE | SVTVEWIENG | DTKGKEIDLE | SIFSLNPDLV | PDEDIEPSPE | TPPPPTSSAK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VNKIVKNRRT | VASIKNEPPP | RDNRVVGSAR | ARPSQLPEQS | SSAQQNARRK | SNCVKEVEKL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QEKREKRRLQ | QQELREKRAQ | DVDATNPNYE | IMCMIRDFRG | SLDYRPLTTA | DPIDEHRICV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| CVRKRPLNKK | ETQMKDLDVI | TIPSKDVVMV | HEPKQKVDLT | RYLENQTFRF | DYAFDDSAPN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EMVYRFTARP | LVETIFERGM | ATCFAYGQTG | SGKTHTMGGD | FSGKNQDCSK | GIYALAARDV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| FLMLKKPNYK | KLELQVNATF | FEIYSGKVFD | LLNRKTKLRV | LEDGKQQVQV | VGLQEREVKC |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VEDVLKLIDI | GNSCRTSGQT | SANAHSSRSH | AVFQIILRRK | GKLHGKFSLI | DLAGNERGAD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TSSADRQTRL | EGAEINKSLL | AHKECIRALG | RNKPHTPFRA | SKLTQVLRDS | FIGENSRTCM |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IATISPGMAS | CENTLNTLRY | ANRVKELTVD | PTAAGDVRPI | MHHPPNQIDD | LEAQWGVGSS |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PQRDDLKLLC | EQNEEEVSPQ | LFTFHEAVSQ | MVEMEEQVVE | DHRAVFQESI | RWLEDEKALL |
| 610 | 620 | 630 | 640 | 650 | |
| EMTEEVDYDV | DSYATQLEAI | LEQKIDILTE | LRDKVKSFRA | ALQEEEQASK | QINPKRPRAL |