Q2KJA1
Gene name |
SH3GL1 |
Protein name |
Endophilin-A2 |
Names |
Endophilin-2, SH3 domain protein 2B, SH3 domain-containing GRB2-like protein 1 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:512719 |
EC number |
|
Protein Class |
|
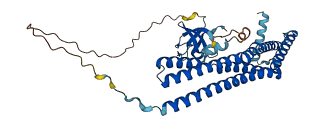
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q2KJA1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q2KJA1-F1 | Predicted | AlphaFoldDB |
152 variants for Q2KJA1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs437349478 | 3 | V>G | No | EVA | |
| rs434861488 | 15 | Q>E | No | EVA | |
| rs444519873 | 47 | V>E | No | EVA | |
| rs453941154 | 63 | A>D | No | EVA | |
| rs472260208 | 66 | A>T | No | EVA | |
| rs442540787 | 67 | K>Q | No | EVA | |
| rs454496033 | 74 | V>A | No | EVA | |
| rs476221466 | 77 | I>S | No | EVA | |
| rs443904223 | 85 | G>R | No | EVA | |
| rs459976978 | 90 | E>D | No | EVA | |
| rs441373156 | 90 | E>G | No | EVA | |
| rs477778798 | 90 | E>Q | No | EVA | |
| rs481546004 | 93 | L>Q | No | EVA | |
| rs448582195 | 95 | E>* | No | EVA | |
| rs469968439 | 98 | I>L | No | EVA | |
| rs482026938 | 100 | H>N | No | EVA | |
| rs446051015 | 101 | G>A | No | EVA | |
| rs432291443 | 103 | E>D | No | EVA | |
| rs465088830 | 103 | E>G | No | EVA | |
| rs466003181 | 105 | G>V | No | EVA | |
| rs454635077 | 108 | S>F | No | EVA | |
| rs435037161 | 112 | D>N | No | EVA | |
| rs475354474 | 121 | M>R | No | EVA | |
| rs442164828 | 123 | R>H | No | EVA | |
| rs442164828 | 123 | R>L | No | EVA | |
| rs463826248 | 124 | L>P | No | EVA | |
| rs475688248 | 134 | E>D | No | EVA | |
| rs458033046 | 147 | C>R | No | EVA | |
| rs447446109 | 150 | D>G | No | EVA | |
| rs481356901 | 152 | K>M | No | EVA | |
| rs448266256 | 152 | K>N | No | EVA | |
| rs481356901 | 152 | K>R | No | EVA | |
| rs469996083 | 155 | Q>K | No | EVA | |
| rs440513822 | 156 | H>L | No | EVA | |
| rs473516314 | 156 | H>N | No | EVA | |
| rs455683696 | 157 | H>N | No | EVA | |
| rs474201780 | 157 | H>P | No | EVA | |
| rs441891338 | 158 | L>V | No | EVA | |
| rs482156013 | 160 | K>R | No | EVA | |
| rs439369887 | 161 | L>R | No | EVA | |
| rs457894095 | 162 | E>G | No | EVA | |
| rs479531685 | 163 | G>R | No | EVA | |
| rs446522707 | 164 | R>P | No | EVA | |
| rs480022288 | 169 | D>A | No | EVA | |
| rs480022288 | 169 | D>G | No | EVA | |
| rs450446010 | 170 | Y>C | No | EVA | |
| rs450446010 | 170 | Y>S | No | EVA | |
| rs433507368 | 173 | K>T | No | EVA | |
| rs434027822 | 177 | K>E | No | EVA | |
| rs434027822 | 177 | K>Q | No | EVA | |
| rs456563204 | 178 | I>M | No | EVA | |
| rs444435493 | 178 | I>N | No | EVA | |
| rs474342590 | 178 | I>V | No | EVA | |
| rs439510499 | 180 | D>A | No | EVA | |
| rs475677633 | 180 | D>H | No | EVA | |
| rs457909050 | 181 | E>Q | No | EVA | |
| rs473194652 | 182 | E>A | No | EVA | |
| rs473194652 | 182 | E>G | No | EVA | |
| rs461677623 | 183 | L>P | No | EVA | |
| rs461677623 | 183 | L>R | No | EVA | |
| rs440043028 | 183 | L>V | No | EVA | |
| rs480163055 | 184 | R>G | No | EVA | |
| rs450384615 | 184 | R>P | No | EVA | |
| rs445496857 | 185 | Q>H | No | EVA | |
| rs478516101 | 185 | Q>P | No | EVA | |
| rs467139857 | 186 | A>G | No | EVA | |
| rs434171337 | 187 | M>L | No | EVA | |
| rs449542625 | 187 | M>T | No | EVA | |
| rs467854281 | 188 | E>A | No | EVA | |
| rs467854281 | 188 | E>G | No | EVA | |
| rs438134269 | 189 | K>E | No | EVA | |
| rs456501564 | 189 | K>T | No | EVA | |
| rs471788726 | 190 | F>V | No | EVA | |
| rs433030945 | 191 | E>* | No | EVA | |
| rs451503386 | 191 | E>G | No | EVA | |
| rs473377639 | 192 | E>G | No | EVA | |
| rs440186577 | 193 | S>A | No | EVA | |
| rs461814380 | 193 | S>F | No | EVA | |
| rs473684413 | 194 | K>Q | No | EVA | |
| rs444123563 | 194 | K>T | No | EVA | |
| rs462338632 | 195 | E>G | No | EVA | |
| rs477857818 | 196 | V>G | No | EVA | |
| rs444874053 | 197 | A>G | No | EVA | |
| rs460787429 | 198 | E>V | No | EVA | |
| rs479392586 | 199 | T>A | No | EVA | |
| rs449478385 | 199 | T>N | No | EVA | |
| rs438071074 | 200 | S>R | No | EVA | |
| rs467993908 | 200 | S>R | No | EVA | |
| rs465324824 | 202 | H>P | No | EVA | |
| rs450222231 | 202 | H>Y | No | EVA | |
| rs432372685 | 203 | H>P | No | EVA | |
| rs432372685 | 203 | H>R | No | EVA | |
| rs454145061 | 204 | L>P | No | EVA | |
| rs473272136 | 206 | E>D | No | EVA | |
| rs433883797 | 207 | T>A | No | EVA | |
| rs455347566 | 207 | T>N | No | EVA | |
| rs455347566 | 207 | T>S | No | EVA | |
| rs473809464 | 208 | D>H | No | EVA | |
| rs473809464 | 208 | D>Y | No | EVA | |
| rs441162956 | 212 | V>G | No | EVA | |
| rs477303127 | 212 | V>L | No | EVA | |
| rs459492496 | 213 | S>G | No | EVA | |
| rs481220606 | 216 | S>P | No | EVA | |
| rs448340041 | 217 | A>D | No | EVA | |
| rs469879354 | 219 | V>A | No | EVA | |
| rs469879354 | 219 | V>E | No | EVA | |
| rs482011978 | 220 | D>N | No | EVA | |
| rs445591600 | 220 | D>V | No | EVA | |
| rs464145907 | 221 | A>P | No | EVA | |
| rs435028746 | 221 | A>V | No | EVA | |
| rs435748858 | 225 | Y>C | No | EVA | |
| rs468747263 | 225 | Y>H | No | EVA | |
| rs435748858 | 225 | Y>S | No | EVA | |
| rs457402788 | 226 | H>R | No | EVA | |
| rs475935330 | 230 | V>G | No | EVA | |
| rs458033138 | 230 | V>L | No | EVA | |
| rs439703377 | 231 | Q>H | No | EVA | |
| rs473388867 | 235 | E>V | No | EVA | |
| rs441100715 | 239 | K>Q | No | EVA | |
| rs459627242 | 241 | K>T | No | EVA | |
| rs481344765 | 243 | R>G | No | EVA | |
| rs477427344 | 244 | M>K | No | EVA | |
| rs458840558 | 244 | M>L | No | EVA | |
| rs458840558 | 244 | M>V | No | EVA | |
| rs448971096 | 256 | K>T | No | EVA | |
| rs467597543 | 258 | K>T | No | EVA | |
| rs456206947 | 262 | L>R | No | EVA | |
| rs431931849 | 264 | D>H | No | EVA | |
| rs453759908 | 265 | L>P | No | EVA | |
| rs461460247 | 267 | E>D | No | EVA | |
| rs443165174 | 267 | E>G | No | EVA | |
| rs443832798 | 271 | S>A | No | EVA | |
| rs459042556 | 271 | S>F | No | EVA | |
| rs477579838 | 274 | G>V | No | EVA | |
| rs447579989 | 275 | F>Y | No | EVA | |
| rs478668524 | 280 | A>P | No | EVA | |
| rs449108186 | 281 | P>T | No | EVA | |
| rs445630407 | 297 | R>Q | No | EVA | |
| rs435068962 | 305 | P>A | No | EVA | |
| rs446974230 | 306 | L>R | No | EVA | |
| rs468608603 | 307 | D>A | No | EVA | |
| rs435635699 | 311 | C>R | No | EVA | |
| rs457358737 | 312 | K>N | No | EVA | |
| rs475996946 | 318 | E>A | No | EVA | |
| rs433152616 | 322 | D>G | No | EVA | |
| rs451755520 | 327 | F>C | No | EVA | |
| rs470845140 | 334 | T>R | No | EVA | |
| rs459492663 | 337 | N>D | No | EVA | |
| rs474802556 | 337 | N>K | No | EVA | |
| rs441855077 | 349 | D>V | No | EVA | |
| rs445571834 | 362 | V>G | No | EVA | |
| rs457600689 | 364 | V>G | No | EVA |
No associated diseases with Q2KJA1
Functions
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| anchoring junction | A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix. |
| cell projection | A prolongation or process extending from a cell, e.g. a flagellum or axon. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early endosome membrane | The lipid bilayer surrounding an early endosome. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| podosome | An actin-rich adhesion structure characterized by formation upon cell substrate contact and localization at the substrate-attached part of the cell, contain an F-actin-rich core surrounded by a ring structure containing proteins such as vinculin and talin, and have a diameter of 0.5 mm. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| lipid binding | Binding to a lipid. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| synaptic vesicle uncoating | The removal of the protein coat on a synaptic vesicle following the pinching step at the end of budding from the presynaptic membrane. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8AXU9 | SH3GL3 | Endophilin-A3 | Gallus gallus (Chicken) | PR |
| Q8AXV1 | SH3GL2 | Endophilin-A1 | Gallus gallus (Chicken) | PR |
| Q99963 | SH3GL3 | Endophilin-A3 | Homo sapiens (Human) | PR |
| Q99962 | SH3GL2 | Endophilin-A1 | Homo sapiens (Human) | PR |
| Q99961 | SH3GL1 | Endophilin-A2 | Homo sapiens (Human) | PR |
| Q9Y371 | SH3GLB1 | Endophilin-B1 | Homo sapiens (Human) | PR |
| Q62421 | Sh3gl3 | Endophilin-A3 | Mus musculus (Mouse) | PR |
| Q62419 | Sh3gl1 | Endophilin-A2 | Mus musculus (Mouse) | PR |
| Q62420 | Sh3gl2 | Endophilin-A1 | Mus musculus (Mouse) | PR |
| Q9JK48 | Sh3glb1 | Endophilin-B1 | Mus musculus (Mouse) | PR |
| O35180 | Sh3gl3 | Endophilin-A3 | Rattus norvegicus (Rat) | PR |
| O35179 | Sh3gl2 | Endophilin-A1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSVAGLKKQF | YKASQLVSEK | VGGAEGTKLD | DDFKEMEKKV | DVTSKAVTEV | LARTIEYLQP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NPASRAKLTM | LNTVSKIRGQ | VKNPGYPQSE | GLLGECMIRH | GKELGGESNF | GDALLDAGES |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MKRLAEVKDS | LDIEVKQNFI | DPLQNLCDKD | LKEIQHHLKK | LEGRRLDFDY | KKKRQGKIPD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EELRQAMEKF | EESKEVAETS | MHHLLETDIE | QVSQLSALVD | AQLDYHRQAV | QILDELADKL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KRRMREASSR | PKREYKPKPR | ELLDLGEPEQ | SNGGFPCAAA | PKITASSSFR | SSDKPVRTPS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RSMPPLDQPS | CKALYDFEPE | NDGELGFHEG | DIITLTNQID | ENWYEGMLDG | QSGFFPLSYV |
| EVLVPLPQ |