Q2HXK9
Gene name |
RBOHD |
Protein name |
Respiratory burst oxidase homolog protein D |
Names |
NADPH oxidase RBOHD, StRBOHD |
Species |
Solanum tuberosum (Potato) |
KEGG Pathway |
sot:102586476 |
EC number |
|
Protein Class |
|
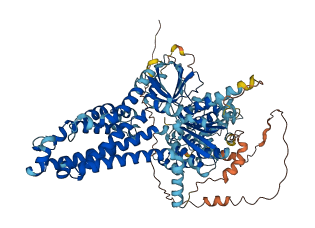
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q2HXK9
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q2HXK9-F1 | Predicted | AlphaFoldDB |
No variants for Q2HXK9
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q2HXK9 | |||||
No associated diseases with Q2HXK9
7 regional properties for Q2HXK9
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EF-hand domain | 175 - 229 | IPR002048 |
| domain | FAD-binding 8 | 557 - 667 | IPR013112 |
| domain | Ferric reductase, NAD binding domain | 674 - 839 | IPR013121 |
| domain | Ferric reductase transmembrane component-like domain | 358 - 514 | IPR013130 |
| domain | NADPH oxidase Respiratory burst | 97 - 197 | IPR013623 |
| domain | FAD-binding domain, ferredoxin reductase-type | 548 - 670 | IPR017927 |
| binding_site | EF-Hand 1, calcium-binding site | 207 - 219 | IPR018247 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| ferric-chelate reductase activity | Catalysis of the reaction: 2 Fe3+-siderophore + electron donor -> 2 Fe3+-siderophore + electron acceptor. |
| NAD(P)H oxidase H2O2-forming activity | Catalysis of the reaction: NAD(P)H + H+ + O2 = NAD(P)+ + hydrogen peroxide. |
| peroxidase activity | Catalysis of the reaction: a donor + a peroxide = an oxidized donor + 2 H2O. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q96PH1 | NOX5 | NADPH oxidase 5 | Homo sapiens (Human) | EV |
| Q948U0 | RBOHA | Respiratory burst oxidase homolog protein A | Solanum tuberosum (Potato) | PR |
| Q948T9 | RBOHB | Respiratory burst oxidase homolog protein B | Solanum tuberosum (Potato) | PR |
| Q9FIJ0 | RBOHD | Respiratory burst oxidase homolog protein D | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MQNPEDHHSD | RELSSPSNTT | KSNDDKNVEI | TLDIRDDTMA | GQSVKNATKT | KAEEAELEAL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GKNLQKKCSF | GATIVRNVSM | RMRLPSFKRQ | PHPPQTFDRS | STAAQNALKG | FKFISKTDGG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SGWDTVQQRF | DELTATSDSL | LPRAKFGECI | GMNRESEGFA | LELFNALARR | RNITSGCISK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EQLKEFWDQI | ANQSFDSRLR | TFFDMVDKDA | DGRLTEEEVR | EIICLSASAN | KLSNIQKQAA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EYAALIMEEL | DRDQKGYIML | ENLEMLLLEA | PIQPDGEKGL | NRNLSHMLSM | KLKPTLETNP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IKRWYNNLKY | FLLDNWRRVW | VLLLWIGVMA | GLFAYKYVQY | KNKAAFNVMG | HCVCVAKGAA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EVLKLNMALI | LLPVCRNTIT | WLRNKTKLGG | AVPFDDNINF | HKVVAGAIAV | GVGIHVLAHM |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TCDFPRLLNA | SPEKYKPMEP | YFGDQPRNYW | HFVKGVEGVS | GIIMVVLMSI | AFTLASQRFR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RNKIRLPRPL | NKLTGFNAFW | YSHHLFVIVY | SLLIVHGIEL | YLTKEWYKKT | TWMYLAIPII |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LYSGERLLRA | FRSSVKDVKI | LKVAMYTGNV | LTLQMSKPQG | FNYKSGQYMF | VNCAAVSPFE |
| 610 | 620 | 630 | 640 | 650 | 660 |
| WHPFSITSAP | GDEYLSVHIR | IVGDWTTKLR | DVFSEPSPTG | RSGLVADYLQ | DKINYPKVLI |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DGPYGAPAQD | YKEYEVLLLV | GLGIGATPMI | SIVKDIVNNM | KEEKYDHDLE | KKTVSGSGRS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| NFKRVYFYWV | TREQGSFDWF | KGLMNELAVM | DCDGIIEMHN | YCTSVYEEGD | ARSALIAMLQ |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SINHAKNGVD | IVSGTRVKTH | FARPNWRNVY | KRIALNHTDA | RVGVFYCGAP | ALTKVLGQLA |
| 850 | |||||
| LDFSHKTSTK | FDFHKENF |