Q1RMR4
Gene name |
RAB15 |
Protein name |
Ras-related protein Rab-15 |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:614507 |
EC number |
|
Protein Class |
|
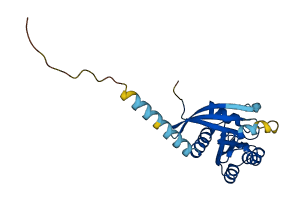
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q1RMR4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q1RMR4-F1 | Predicted | AlphaFoldDB |
61 variants for Q1RMR4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs43638279 | 5 | Y>C | No | EVA | |
| rs447981776 | 5 | Y>D | No | EVA | |
| rs43638279 | 5 | Y>S | No | EVA | |
| rs483113706 | 7 | V>G | No | EVA | |
| rs450354872 | 7 | V>L | No | EVA | |
| rs441097469 | 11 | L>M | No | EVA | |
| rs443435879 | 15 | G>R | No | EVA | |
| rs435956732 | 16 | D>E | No | EVA | |
| rs454490521 | 16 | D>G | No | EVA | |
| rs476095802 | 16 | D>Y | No | EVA | |
| rs465780342 | 17 | S>C | No | EVA | |
| rs465780342 | 17 | S>Y | No | EVA | |
| rs432650880 | 19 | V>G | No | EVA | |
| rs483235174 | 22 | T>A | No | EVA | |
| rs467837625 | 22 | T>I | No | EVA | |
| rs483235174 | 22 | T>P | No | EVA | |
| rs479002859 | 23 | C>* | No | EVA | |
| rs460564428 | 25 | L>V | No | EVA | |
| rs438825733 | 26 | C>S | No | EVA | |
| rs458708012 | 27 | R>G | No | EVA | |
| rs443372373 | 28 | F>S | No | EVA | |
| rs476234891 | 29 | T>P | No | EVA | |
| rs454427273 | 30 | D>A | No | EVA | |
| rs442515045 | 30 | D>E | No | EVA | |
| rs454427273 | 30 | D>G | No | EVA | |
| rs472318820 | 32 | E>G | No | EVA | |
| rs453680742 | 34 | H>D | No | EVA | |
| rs431989636 | 34 | H>L | No | EVA | |
| rs431989636 | 34 | H>P | No | EVA | |
| rs456843145 | 35 | S>P | No | EVA | |
| rs438179357 | 36 | S>L | No | EVA | |
| rs449250704 | 37 | H>L | No | EVA | |
| rs449250704 | 37 | H>P | No | EVA | |
| rs467086882 | 38 | I>T | No | EVA | |
| rs445209124 | 40 | T>P | No | EVA | |
| rs478288890 | 41 | I>M | No | EVA | |
| rs454019504 | 68 | E>G | No | EVA | |
| rs435843043 | 94 | R>C | No | EVA | |
| rs468714802 | 94 | R>H | No | EVA | |
| rs446993656 | 96 | Y>H | No | EVA | |
| rs446084222 | 99 | I>V | No | EVA | |
| rs385478748 | 107 | D>E | No | EVA | |
| rs436319121 | 131 | V>G | No | EVA | |
| rs467887392 | 148 | Y>* | No | EVA | |
| rs446881358 | 154 | T>P | No | EVA | |
| rs475883540 | 163 | T>P | No | EVA | |
| rs386132009 | 163 | T>R | No | EVA | |
| rs468747815 | 174 | R>S | No | EVA | |
| rs434839150 | 179 | G>A | No | EVA | |
| rs434839150 | 179 | G>V | No | EVA | |
| rs470209470 | 187 | E>D | No | EVA | |
| rs450673490 | 188 | L>* | No | EVA | |
| rs480594779 | 191 | A>T | No | EVA | |
| rs461907660 | 192 | E>G | No | EVA | |
| rs479265329 | 198 | G>S | No | EVA | |
| rs457700815 | 198 | G>V | No | EVA | |
| rs439127840 | 199 | K>E | No | EVA | |
| rs475185695 | 201 | E>A | No | EVA | |
| rs210569353 | 203 | P>L | No | EVA | |
| rs456968991 | 209 | T>P | No | EVA | |
| rs442282734 | 211 | W>S | No | EVA |
No associated diseases with Q1RMR4
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| cilium | A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface and of some cytoplasmic parts. Each cilium is largely bounded by an extrusion of the cytoplasmic (plasma) membrane, and contains a regular longitudinal array of microtubules, anchored to a basal body. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| insulin-responsive compartment | A small membrane-bounded vesicle that releases its contents by exocytosis in response to insulin stimulation; the contents are enriched in GLUT4, IRAP and VAMP2. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
| synaptic vesicle | A secretory organelle, typically 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and secretes these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| positive regulation of regulated secretory pathway | Any process that activates or increases the frequency, rate or extent of regulated secretory pathway. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein secretion | The controlled release of proteins from a cell. |
| Rab protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rab family of proteins switching to a GTP-bound active state. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| vesicle docking involved in exocytosis | The initial attachment of a vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the vesicle and the target membrane, that contributes to exocytosis. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A4FV54 | RAB8A | Ras-related protein Rab-8A | Bos taurus (Bovine) | PR |
| Q2HJI8 | RAB8B | Ras-related protein Rab-8B | Bos taurus (Bovine) | PR |
| Q5F470 | RAB8A | Ras-related protein Rab-8A | Gallus gallus (Chicken) | PR |
| P0C0E4 | RAB40AL | Ras-related protein Rab-40A-like | Homo sapiens (Human) | PR |
| P61006 | RAB8A | Ras-related protein Rab-8A | Homo sapiens (Human) | PR |
| Q92930 | RAB8B | Ras-related protein Rab-8B | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q8WXH6 | RAB40A | Ras-related protein Rab-40A | Homo sapiens (Human) | PR |
| Q96S21 | RAB40C | Ras-related protein Rab-40C | Homo sapiens (Human) | PR |
| Q12829 | RAB40B | Ras-related protein Rab-40B | Homo sapiens (Human) | PR |
| P59190 | RAB15 | Ras-related protein Rab-15 | Homo sapiens (Human) | PR |
| P55258 | Rab8a | Ras-related protein Rab-8A | Mus musculus (Mouse) | PR |
| P61028 | Rab8b | Ras-related protein Rab-8B | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q9DD03 | Rab13 | Ras-related protein Rab-13 | Mus musculus (Mouse) | PR |
| Q8CB87 | Rab44 | Ras-related protein Rab-44 | Mus musculus (Mouse) | PR |
| Q8VHP8 | Rab40b | Ras-related protein Rab-40B | Mus musculus (Mouse) | PR |
| Q8VHQ4 | Rab40c | Ras-related protein Rab-40C | Mus musculus (Mouse) | PR |
| Q8K386 | Rab15 | Ras-related protein Rab-15 | Mus musculus (Mouse) | PR |
| P35280 | Rab8a | Ras-related protein Rab-8A | Rattus norvegicus (Rat) | PR |
| P70550 | Rab8b | Ras-related protein Rab-8B | Rattus norvegicus (Rat) | PR |
| P35284 | Rab12 | Ras-related protein Rab-12 | Rattus norvegicus (Rat) | PR |
| P35281 | Rab10 | Ras-related protein Rab-10 | Rattus norvegicus (Rat) | PR |
| P35289 | Rab15 | Ras-related protein Rab-15 | Rattus norvegicus (Rat) | PR |
| Q9SF91 | RABE1E | Ras-related protein RABE1e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O24466 | RABE1A | Ras-related protein RABE1a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LZD4 | RABE1D | Ras-related protein RABE1d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAKQYDVLFR | LLLIGDSGVG | KTCLLCRFTD | NEFHSSHIST | IGVDFKMKTI | EVDGIKVRIQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IWDTAGQERY | QTITKQYYRR | AQGIFLVYDI | SSERSYQHIM | KWVSDVDEYA | PEGVQKILIG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NKADEEQKRQ | VGREQGQQLA | REYGMDFYET | SACTNLNIKE | SFTRLTELVL | QAHRKELEGL |
| 190 | 200 | 210 | |||
| RTRANHELAL | AELEEDEGKP | EGPANSSKTC | WC |