Q1RML2
Gene name |
PLCZ1 |
Protein name |
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 |
Names |
Phosphoinositide phospholipase C-zeta-1, Phospholipase C-zeta-1, PLC-zeta-1 |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:497026 |
EC number |
3.1.4.11: Phosphoric diester hydrolases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
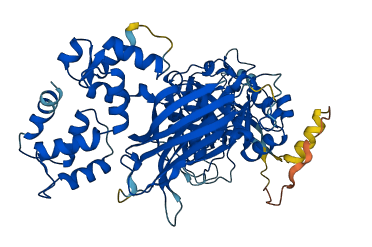
Autoinhibited structure

Activated structure

1 structures for Q1RML2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q1RML2-F1 | Predicted | AlphaFoldDB |
87 variants for Q1RML2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs479021129 | 41 | K>E | No | EVA | |
| rs434350358 | 57 | I>L | No | EVA | |
| rs452949377 | 63 | I>L | No | EVA | |
| rs452949377 | 63 | I>V | No | EVA | |
| rs465039260 | 76 | I>N | No | EVA | |
| rs432091682 | 90 | K>E | No | EVA | |
| rs451784478 | 96 | L>W | No | EVA | |
| rs476842731 | 100 | Q>* | No | EVA | |
| rs443932700 | 102 | T>I | No | EVA | |
| rs456036670 | 103 | L>F | No | EVA | |
| rs474148086 | 105 | F>L | No | EVA | |
| rs441265316 | 109 | I>F | No | EVA | |
| rs441265316 | 109 | I>L | No | EVA | |
| rs459815659 | 109 | I>N | No | EVA | |
| rs478337847 | 110 | A>P | No | EVA | |
| rs438916541 | 110 | A>V | No | EVA | |
| rs463518431 | 111 | S>T | No | EVA | |
| rs449048927 | 114 | I>N | No | EVA | |
| rs467523606 | 115 | Q>K | No | EVA | |
| rs479177598 | 115 | Q>L | No | EVA | |
| rs479177598 | 115 | Q>R | No | EVA | |
| rs432057147 | 117 | Y>N | No | EVA | |
| rs457063279 | 120 | I>V | No | EVA | |
| rs457011048 | 255 | S>C | No | EVA | |
| rs468329045 | 256 | Q>* | No | EVA | |
| rs797852517 | 267 | T>I | No | EVA | |
| rs445141274 | 321 | E>G | No | EVA | |
| rs469572073 | 322 | F>S | No | EVA | |
| rs208019489 | 337 | A>P | No | EVA | |
| rs716682782 | 361 | G>R | No | EVA | |
| rs470423282 | 370 | I>K | No | EVA | |
| rs437484228 | 372 | I>K | No | EVA | |
| rs456024945 | 373 | S>F | No | EVA | |
| rs434799806 | 374 | M>I | No | EVA | |
| rs474162142 | 374 | M>L | No | EVA | |
| rs453306797 | 375 | A>P | No | EVA | |
| rs471695736 | 376 | L>P | No | EVA | |
| rs463383010 | 377 | S>F | No | EVA | |
| rs475415030 | 378 | D>V | No | EVA | |
| rs442644313 | 379 | L>F | No | EVA | |
| rs479415700 | 380 | V>F | No | EVA | |
| rs446418300 | 381 | I>F | No | EVA | |
| rs458504986 | 382 | Y>F | No | EVA | |
| rs477095828 | 390 | S>R | No | EVA | |
| rs450657575 | 404 | N>Y | No | EVA | |
| rs470339921 | 410 | Q>K | No | EVA | |
| rs449472003 | 414 | L>I | No | EVA | |
| rs468108775 | 417 | L>* | No | EVA | |
| rs435115031 | 417 | L>F | No | EVA | |
| rs437891986 | 421 | E>K | No | EVA | |
| rs456440454 | 422 | F>Y | No | EVA | |
| rs474577186 | 423 | I>T | No | EVA | |
| rs435192504 | 425 | H>N | No | EVA | |
| rs472249847 | 429 | F>S | No | EVA | |
| rs453723767 | 429 | F>V | No | EVA | |
| rs458970104 | 430 | I>T | No | EVA | |
| rs439307697 | 430 | I>V | No | EVA | |
| rs463238307 | 432 | R>S | No | EVA | |
| rs481430307 | 433 | V>E | No | EVA | |
| rs479105501 | 434 | Y>F | No | EVA | |
| rs460582806 | 434 | Y>H | No | EVA | |
| rs446205489 | 436 | K>E | No | EVA | |
| rs464317848 | 436 | K>T | No | EVA | |
| rs482820740 | 437 | A>G | No | EVA | |
| rs449930011 | 444 | N>I | No | EVA | |
| rs468427947 | 446 | N>H | No | EVA | |
| rs435500869 | 446 | N>T | No | EVA | |
| rs465819127 | 448 | Q>E | No | EVA | |
| rs451301128 | 449 | E>D | No | EVA | |
| rs432806001 | 449 | E>Q | No | EVA | |
| rs470979464 | 450 | F>V | No | EVA | |
| rs444616223 | 451 | W>* | No | EVA | |
| rs456754408 | 452 | N>I | No | EVA | |
| rs460495241 | 456 | Q>* | No | EVA | |
| rs479019352 | 457 | M>L | No | EVA | |
| rs382574994 | 489 | R>G | No | EVA | |
| rs433503815 | 496 | T>I | No | EVA | |
| rs476610966 | 497 | K>E | No | EVA | |
| rs473657495 | 586 | G>D | No | EVA | |
| rs482888544 | 608 | Y>S | No | EVA | |
| rs449866686 | 609 | R>H | No | EVA | |
| rs449866686 | 609 | R>P | No | EVA | |
| rs447198976 | 617 | M>I | No | EVA | |
| rs462037939 | 617 | M>L | No | EVA | |
| rs480347282 | 617 | M>R | No | EVA | |
| rs432805987 | 618 | G>A | No | EVA | |
| rs465638405 | 618 | G>C | No | EVA |
No associated diseases with Q1RML2
5 regional properties for Q1RML2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 492 - 615 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 155 - 300 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 376 - 492 | IPR001711 |
| domain | EF-hand domain | 35 - 70 | IPR002048 |
| domain | Phosphoinositide-specific phospholipase C, EF-hand-like domain | 64 - 147 | IPR015359 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.11 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| pronucleus | The nucleus of either the ovum or the spermatozoon following fertilization. Thus, in the fertilized ovum, there are two pronuclei, one originating from the ovum, the other from the spermatozoon that brought about fertilization; they approach each other, but do not fuse until just before the first cleavage, when each pronucleus loses its membrane to release its contents. |
| sperm head | The part of the late spermatid or spermatozoon that contains the nucleus and acrosome. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| phosphatidylinositol phospholipase C activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H(2)O = 1,2-diacylglycerol + 1D-myo-inositol 1,4,5-trisphosphate + H(+). |
| phosphatidylinositol-3-phosphate binding | Binding to phosphatidylinositol-3-phosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 3' position. |
| phosphatidylinositol-4,5-bisphosphate binding | Binding to phosphatidylinositol-4,5-bisphosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 4' and 5' positions. |
| phosphatidylinositol-5-phosphate binding | Binding to phosphatidylinositol-5-phosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 5' position. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| calcium ion transport | The directed movement of calcium (Ca) ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| egg activation | The process in which the egg becomes metabolically active, initiates protein and DNA synthesis and undergoes structural changes to its cortex and/or cytoplasm. |
| lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| positive regulation of cytosolic calcium ion concentration involved in egg activation | The process that increases the concentration of calcium ions in the cytosol after fertilization or the physiological activation of an egg. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32383 | PLC1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P10895 | PLCD1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Bos taurus (Bovine) | SS |
| P10894 | PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Bos taurus (Bovine) | SS |
| P08487 | PLCG1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Bos taurus (Bovine) | EV SS |
| Q2VRL0 | PLCZ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Gallus gallus (Chicken) | PR |
| Q9P212 | PLCE1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Homo sapiens (Human) | SS |
| Q9BRC7 | PLCD4 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-4 | Homo sapiens (Human) | SS |
| Q15111 | PLCL1 | Inactive phospholipase C-like protein 1 | Homo sapiens (Human) | PR |
| P51178 | PLCD1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Homo sapiens (Human) | EV |
| Q9UPR0 | PLCL2 | Inactive phospholipase C-like protein 2 | Homo sapiens (Human) | PR |
| Q8N3E9 | PLCD3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | Homo sapiens (Human) | SS |
| Q86YW0 | PLCZ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Homo sapiens (Human) | PR |
| Q8R3B1 | Plcd1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Mus musculus (Mouse) | SS |
| Q8K394 | Plcl2 | Inactive phospholipase C-like protein 2 | Mus musculus (Mouse) | PR |
| Q8K2J0 | Plcd3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | Mus musculus (Mouse) | PR |
| Q8K4S1 | Plce1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Mus musculus (Mouse) | SS |
| Q7YRU3 | PLCZ | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 | Sus scrofa (Pig) | PR |
| P10688 | Plcd1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Rattus norvegicus (Rat) | SS |
| Q99P84 | Plce1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Rattus norvegicus (Rat) | EV |
| Q39032 | PLC1 | Phosphoinositide phospholipase C 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q56W08 | PLC3 | Phosphoinositide phospholipase C 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NMA7 | PLC9 | Phosphoinositide phospholipase C 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8GV43 | PLC6 | Phosphoinositide phospholipase C 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C2 | PLC5 | Phosphoinositide phospholipase C 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STZ3 | PLC8 | Phosphoinositide phospholipase C 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C1 | PLC4 | Phosphoinositide phospholipase C 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| A5D6R3 | plcd3a | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3-A | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MENKWFLLMV | RDDFKGGKIT | LEKALKLLEK | LDIQCNTIHV | KYIFKDNDRL | KQGRITIEEF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RTIYRIITYR | EEIIEIFNTY | SENRKILLEK | NLVEFLMREQ | YTLDFNKSIA | SEIIQKYEPI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EEVKQAHQMS | FEGFRRYMDS | SECLLFDNKC | DHVYQDMTHP | LTDYFISSSH | NTYLISDQLW |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GPSDLWGYIS | ALVKGCRCLE | IDCWDGSQNE | PVVYHGYTFT | SKLLFKTVIQ | AINKYAFLAS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EYPVVLSLEN | HCSPSQQEVM | ADSLLATFGD | ALLSYTLDNF | SDRLPSPEAL | KFKILVRNKK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IGTLHETLER | KGSDMHGKVE | EFEEEEEIEQ | EEDGSGAKEP | EPVGDFQDDL | AKEEQLKRVV |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GIPLFRKKKI | KISMALSDLV | IYTKVEKFKS | FHHSHLYQQF | NESNSIGESQ | ARKLTKLAAR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EFILHTRRFI | TRVYPKALRA | DSSNFNPQEF | WNVGCQMVAL | NFQTPGVPMD | LQNGKFLDNG |
| 490 | 500 | 510 | 520 | 530 | 540 |
| CSGYVLKPRF | LRDKKTKFNP | HKVQIDSNPL | TLTIRLISGI | QLPPSYQNKA | DTLVIVEIFG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VPNDQMKQQS | RVIKKNAFNP | RWNETFTFVI | QVPELALIRF | VAENQGLIAG | NEFLGQYTLP |
| 610 | 620 | 630 | |||
| VLCMNRGYRR | VPLFSKMGES | LEPASLFIYV | WYIR |