Q1PEC0
Gene name |
PME42 (ARATH42, At4g03930, T24M8.6) |
Protein name |
Probable pectinesterase/pectinesterase inhibitor 42 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G03930 |
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
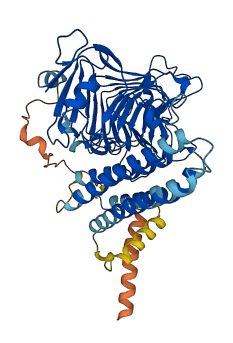
Descriptions
(Annotation from UniProt)
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
215-509 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q1PEC0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q1PEC0-F1 | Predicted | AlphaFoldDB |
265 variants for Q1PEC0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_4_1870488_G_A | 23 | E>K | No | 1000Genomes | |
| ENSVATH02641502 | 29 | H>Q | No | 1000Genomes | |
| ENSVATH02641502 | 29 | H>Q | No | 1000Genomes | |
| ENSVATH10642468 | 31 | C>Y | No | 1000Genomes | |
| ENSVATH13909320 | 40 | T>I | No | 1000Genomes | |
| ENSVATH13909321 | 41 | A>T | No | 1000Genomes | |
| ENSVATH08219187 | 42 | S>A | No | 1000Genomes | |
| ENSVATH13909322 | 43 | S>L | No | 1000Genomes | |
| ENSVATH02641504 | 44 | S>* | No | 1000Genomes | |
| ENSVATH06446267 | 46 | S>A | No | 1000Genomes | |
| ENSVATH10642470 | 49 | R>K | No | 1000Genomes | |
| ENSVATH02641505 | 51 | R>C | No | 1000Genomes | |
| ENSVATH06446269 | 51 | R>H | No | 1000Genomes | |
| ENSVATH02641506 | 52 | D>N | No | 1000Genomes | |
| tmp_4_1870590_G_A | 57 | V>I | No | 1000Genomes | |
| ENSVATH06446273 | 60 | N>K | No | 1000Genomes | |
| ENSVATH06446274 | 61 | S>F | No | 1000Genomes | |
| ENSVATH13909323 | 62 | V>M | No | 1000Genomes | |
| ENSVATH02641510 | 63 | R>Q | No | 1000Genomes | |
| ENSVATH02641509 | 63 | R>W | No | 1000Genomes | |
| ENSVATH06446275 | 64 | K>* | No | 1000Genomes | |
| ENSVATH06446276 | 67 | M>I | No | 1000Genomes | |
| tmp_4_1870624_C_T | 68 | A>V | No | 1000Genomes | |
| ENSVATH02641512 | 71 | G>C | No | 1000Genomes | |
| tmp_4_1870643_A_C | 74 | E>D | No | 1000Genomes | |
| ENSVATH02641513 | 75 | D>N | No | 1000Genomes | |
| tmp_4_1870648_C_T | 76 | T>I | No | 1000Genomes | |
| ENSVATH10642473 | 80 | E>D | No | 1000Genomes | |
| tmp_4_1870659_G_A | 80 | E>K | No | 1000Genomes | |
| tmp_4_1870678_A_T | 86 | K>M | No | 1000Genomes | |
| ENSVATH06446277 | 88 | C>* | No | 1000Genomes | |
| ENSVATH10642495 | 88 | C>G | No | 1000Genomes | |
| ENSVATH02641516 | 89 | L>F | No | 1000Genomes | |
| tmp_4_1870694_G_T | 91 | E>D | No | 1000Genomes | |
| ENSVATH10642497 | 96 | F>L | No | 1000Genomes | |
| ENSVATH08219188 | 97 | E>G | No | 1000Genomes | |
| tmp_4_1870710_G_C | 97 | E>Q | No | 1000Genomes | |
| ENSVATH02641517 | 102 | S>A | No | 1000Genomes | |
| tmp_4_1870729_C_G | 103 | A>G | No | 1000Genomes | |
| ENSVATH02641519 | 107 | M>I | No | 1000Genomes | |
| ENSVATH10642498 | 108 | I>N | No | 1000Genomes | |
| tmp_4_1870746_C_T | 109 | R>C | No | 1000Genomes | |
| ENSVATH06446279 | 109 | R>H | No | 1000Genomes | |
| ENSVATH02641520 | 110 | S>L | No | 1000Genomes | |
| tmp_4_1870752_G_A | 111 | V>M | No | 1000Genomes | |
| ENSVATH08219190 | 115 | L>S | No | 1000Genomes | |
| tmp_4_1870764_T_G | 115 | L>V | No | 1000Genomes | |
| ENSVATH08219191 | 116 | G>* | No | 1000Genomes | |
| tmp_4_1870770_G_A | 117 | G>R | No | 1000Genomes | |
| ENSVATH06446280 | 118 | E>Q | No | 1000Genomes | |
| ENSVATH10642499 | 120 | P>L | No | 1000Genomes | |
| ENSVATH06446282 | 121 | Y>C | No | 1000Genomes | |
| ENSVATH08219192 | 122 | L>P | No | 1000Genomes | |
| ENSVATH13909355 | 129 | H>R | No | 1000Genomes | |
| ENSVATH06446283 | 129 | H>Y | No | 1000Genomes | |
| ENSVATH06446284 | 130 | T>N | No | 1000Genomes | |
| tmp_4_1870815_C_T | 132 | L>F | No | 1000Genomes | |
| ENSVATH06446285 | 134 | G>D | No | 1000Genomes | |
| ENSVATH06446286 | 136 | L>F | No | 1000Genomes | |
| tmp_4_1870842_A_G | 141 | T>A | No | 1000Genomes | |
| tmp_4_1870846_G_A | 142 | C>Y | No | 1000Genomes | |
| ENSVATH06446289 | 144 | D>N | No | 1000Genomes | |
| ENSVATH06446289 | 144 | D>Y | No | 1000Genomes | |
| tmp_4_1870867_G_A | 149 | G>D | No | 1000Genomes | |
| ENSVATH06446291 | 149 | G>S | No | 1000Genomes | |
| tmp_4_1870869_G_A | 150 | A>T | No | 1000Genomes | |
| ENSVATH13909357 | 154 | R>W | No | 1000Genomes | |
| tmp_4_1870887_G_A | 156 | E>K | No | 1000Genomes | |
| ENSVATH10642502 | 157 | P>T | No | 1000Genomes | |
| ENSVATH06446292 | 162 | L>F | No | 1000Genomes | |
| tmp_4_1870909_T_A | 163 | I>N | No | 1000Genomes | |
| ENSVATH13909358 | 164 | S>F | No | 1000Genomes | |
| tmp_4_1870914_A_T | 165 | K>* | No | 1000Genomes | |
| ENSVATH06446293 | 165 | K>R | No | 1000Genomes | |
| ENSVATH06446295 | 166 | A>T | No | 1000Genomes | |
| tmp_4_1870930_TG_CG,T | 170 | L>S | No | 1000Genomes | |
| ENSVATH06446296 | 172 | L>R | No | 1000Genomes | |
| ENSVATH06446298 | 177 | S>* | No | 1000Genomes | |
| ENSVATH06446298 | 177 | S>L | No | 1000Genomes | |
| ENSVATH02641525 | 178 | P>L | No | 1000Genomes | |
| ENSVATH02641525 | 178 | P>Q | No | 1000Genomes | |
| ENSVATH10642545 | 179 | R>K | No | 1000Genomes | |
| ENSVATH02641527 | 183 | E>K | No | 1000Genomes | |
| ENSVATH02641528 | 184 | L>F | No | 1000Genomes | |
| ENSVATH06446300 | 186 | S>L | No | 1000Genomes | |
| ENSVATH02641529 | 189 | P>L | No | 1000Genomes | |
| tmp_4_1870986_C_T | 189 | P>S | No | 1000Genomes | |
| ENSVATH08219201 | 191 | S>N | No | 1000Genomes | |
| tmp_4_1871013_G_T | 198 | V>F | No | 1000Genomes | |
| ENSVATH13909359 | 199 | D>E | No | 1000Genomes | |
| tmp_4_1871016_G_T | 199 | D>Y | No | 1000Genomes | |
| ENSVATH06446302 | 202 | D>N | No | 1000Genomes | |
| ENSVATH02641532 | 207 | A>D | No | 1000Genomes | |
| ENSVATH02641532 | 207 | A>G | No | 1000Genomes | |
| ENSVATH02641531 | 207 | A>S | No | 1000Genomes | |
| ENSVATH02641532 | 207 | A>V | No | 1000Genomes | |
| ENSVATH06446304 | 208 | E>K | No | 1000Genomes | |
| ENSVATH06446304 | 208 | E>Q | No | 1000Genomes | |
| ENSVATH10642550 | 209 | A>S | No | 1000Genomes | |
| tmp_4_1871331_C_T | 209 | A>V | No | 1000Genomes | |
| ENSVATH06446332 | 213 | I>V | No | 1000Genomes | |
| ENSVATH02641545 | 214 | A>D | No | 1000Genomes | |
| ENSVATH02641545 | 214 | A>V | No | 1000Genomes | |
| tmp_4_1871349_A_G | 215 | D>G | No | 1000Genomes | |
| tmp_4_1871355_T_C | 217 | V>A | No | 1000Genomes | |
| ENSVATH13909376 | 219 | A>T | No | 1000Genomes | |
| ENSVATH13909377 | 219 | A>V | No | 1000Genomes | |
| tmp_4_1871366_G_T | 221 | D>Y | No | 1000Genomes | |
| tmp_4_1871370_G_A | 222 | G>E | No | 1000Genomes | |
| ENSVATH06446333 | 224 | G>R | No | 1000Genomes | |
| ENSVATH10642551 | 226 | Y>D | No | 1000Genomes | |
| ENSVATH10642552 | 227 | N>S | No | 1000Genomes | |
| tmp_4_1871388_C_T | 228 | T>I | No | 1000Genomes | |
| ENSVATH08219212 | 231 | A>V | No | 1000Genomes | |
| ENSVATH10642554 | 232 | A>T | No | 1000Genomes | |
| ENSVATH02641551 | 234 | A>V | No | 1000Genomes | |
| ENSVATH06446334 | 235 | A>T | No | 1000Genomes | |
| ENSVATH08219213 | 236 | A>T | No | 1000Genomes | |
| tmp_4_1871414_C_A | 237 | P>T | No | 1000Genomes | |
| ENSVATH06446335 | 238 | Q>L | No | 1000Genomes | |
| ENSVATH06446335 | 238 | Q>R | No | 1000Genomes | |
| ENSVATH06446336 | 239 | H>Y | No | 1000Genomes | |
| tmp_4_1871424_G_A | 240 | S>N | No | 1000Genomes | |
| tmp_4_1871428_C_G | 241 | H>Q | No | 1000Genomes | |
| tmp_4_1871430_A_G | 242 | K>R | No | 1000Genomes | |
| ENSVATH10642605 | 248 | I>K | No | 1000Genomes | |
| ENSVATH06446338 | 250 | T>I | No | 1000Genomes | |
| ENSVATH06446338 | 250 | T>R | No | 1000Genomes | |
| ENSVATH02641554 | 251 | G>S | No | 1000Genomes | |
| ENSVATH13909378 | 253 | Y>C | No | 1000Genomes | |
| ENSVATH13909378 | 253 | Y>F | No | 1000Genomes | |
| ENSVATH10642606 | 253 | Y>H | No | 1000Genomes | |
| ENSVATH10642606 | 253 | Y>N | No | 1000Genomes | |
| ENSVATH06446340 | 254 | D>E | No | 1000Genomes | |
| ENSVATH10642607 | 256 | I>T | No | 1000Genomes | |
| ENSVATH13909379 | 256 | I>V | No | 1000Genomes | |
| ENSVATH02641555 | 258 | A>T | No | 1000Genomes | |
| tmp_4_1871483_G_A | 260 | E>K | No | 1000Genomes | |
| ENSVATH13909380 | 262 | T>M | No | 1000Genomes | |
| tmp_4_1871516_G_A | 271 | D>N | No | 1000Genomes | |
| ENSVATH10642608 | 275 | S>* | No | 1000Genomes | |
| tmp_4_1871541_C_T | 279 | T>M | No | 1000Genomes | |
| ENSVATH02641556 | 289 | R>K | No | 1000Genomes | |
| ENSVATH06446345 | 292 | Y>F | No | 1000Genomes | |
| tmp_4_1871583_C_G | 293 | T>S | No | 1000Genomes | |
| ENSVATH06446346 | 294 | A>T | No | 1000Genomes | |
| ENSVATH06446347 | 294 | A>V | No | 1000Genomes | |
| tmp_4_1871589_C_T | 295 | T>I | No | 1000Genomes | |
| tmp_4_1871705_C_A | 298 | S>Y | No | 1000Genomes | |
| ENSVATH13909385 | 299 | N>T | No | 1000Genomes | |
| tmp_4_1871731_G_C | 307 | D>H | No | 1000Genomes | |
| ENSVATH10642614 | 309 | C>F | No | 1000Genomes | |
| tmp_4_1871739_C_G | 309 | C>W | No | 1000Genomes | |
| ENSVATH02641561 | 311 | R>* | No | 1000Genomes | |
| ENSVATH08219215 | 313 | T>A | No | 1000Genomes | |
| ENSVATH06446354 | 313 | T>M | No | 1000Genomes | |
| ENSVATH02641562 | 314 | V>A | No | 1000Genomes | |
| ENSVATH02641564 | 315 | G>E | No | 1000Genomes | |
| ENSVATH02641563 | 315 | G>R | No | 1000Genomes | |
| ENSVATH10642637 | 316 | P>L | No | 1000Genomes | |
| tmp_4_1871761_G_C | 317 | A>P | No | 1000Genomes | |
| tmp_4_1871768_G_A | 319 | G>E | No | 1000Genomes | |
| ENSVATH02641565 | 321 | A>V | No | 1000Genomes | |
| ENSVATH06446355 | 323 | A>T | No | 1000Genomes | |
| ENSVATH13909386 | 327 | S>C | No | 1000Genomes | |
| ENSVATH06446357 | 328 | G>S | No | 1000Genomes | |
| ENSVATH09519468 | 332 | V>I | No | 1000Genomes | |
| ENSVATH06446358 | 336 | C>Y | No | 1000Genomes | |
| ENSVATH02641568 | 338 | V>I | No | 1000Genomes | |
| tmp_4_1871837_A_T | 342 | Q>L | No | 1000Genomes | |
| ENSVATH02641571 | 344 | A>T | No | 1000Genomes | |
| tmp_4_1871847_G_C | 345 | L>F | No | 1000Genomes | |
| ENSVATH06446359 | 347 | P>S | No | 1000Genomes | |
| ENSVATH02641572 | 348 | H>Q | No | 1000Genomes | |
| tmp_4_1871860_G_C | 350 | D>H | No | 1000Genomes | |
| ENSVATH06446360 | 352 | Q>* | No | 1000Genomes | |
| ENSVATH10642641 | 353 | F>L | No | 1000Genomes | |
| tmp_4_1871876_G_A | 355 | R>K | No | 1000Genomes | |
| ENSVATH10642642 | 355 | R>S | No | 1000Genomes | |
| ENSVATH13909387 | 356 | E>* | No | 1000Genomes | |
| ENSVATH13909388 | 357 | C>W | No | 1000Genomes | |
| ENSVATH02641573 | 358 | F>V | No | 1000Genomes | |
| ENSVATH06446361 | 358 | F>Y | No | 1000Genomes | |
| tmp_4_1871894_G_A | 361 | G>D | No | 1000Genomes | |
| tmp_4_1871899_G_A | 363 | V>I | No | 1000Genomes | |
| tmp_4_1871915_G_A | 368 | G>E | No | 1000Genomes | |
| tmp_4_1871920_G_A | 370 | A>T | No | 1000Genomes | |
| tmp_4_1871923_G_C | 371 | A>P | No | 1000Genomes | |
| tmp_4_1871924_C_T | 371 | A>V | No | 1000Genomes | |
| ENSVATH13909389 | 372 | A>T | No | 1000Genomes | |
| tmp_4_1871943_C_G | 377 | C>W | No | 1000Genomes | |
| tmp_4_1871944_C_G | 378 | Q>E | No | 1000Genomes | |
| ENSVATH10642665 | 378 | Q>H | No | 1000Genomes | |
| tmp_4_1871954_C_T | 381 | A>V | No | 1000Genomes | |
| ENSVATH06446365 | 382 | R>K | No | 1000Genomes | |
| ENSVATH06446366 | 383 | Q>* | No | 1000Genomes | |
| ENSVATH06446366 | 383 | Q>K | No | 1000Genomes | |
| ENSVATH02641575 | 385 | N>K | No | 1000Genomes | |
| tmp_4_1871974_C_A | 388 | Q>K | No | 1000Genomes | |
| tmp_4_1871980_A_G | 390 | N>D | No | 1000Genomes | |
| tmp_4_1871981_A_T | 390 | N>I | No | 1000Genomes | |
| ENSVATH02641576 | 391 | F>V | No | 1000Genomes | |
| ENSVATH10642666 | 394 | A>V | No | 1000Genomes | |
| ENSVATH10642667 | 397 | R>H | No | 1000Genomes | |
| ENSVATH06446371 | 402 | D>E | No | 1000Genomes | |
| tmp_4_1872016_G_A | 402 | D>N | No | 1000Genomes | |
| ENSVATH00460010 | 404 | S>L | No | 1000Genomes | |
| ENSVATH02641578 | 405 | G>D | No | 1000Genomes | |
| ENSVATH06446373 | 411 | C>Y | No | 1000Genomes | |
| ENSVATH02641580 | 415 | A>T | No | 1000Genomes | |
| ENSVATH02641581 | 417 | S>L | No | 1000Genomes | |
| ENSVATH10642668 | 418 | D>N | No | 1000Genomes | |
| ENSVATH06446374 | 421 | T>I | No | 1000Genomes | |
| ENSVATH06446375 | 422 | A>G | No | 1000Genomes | |
| ENSVATH02641583 | 422 | A>S | No | 1000Genomes | |
| ENSVATH02641583 | 422 | A>T | No | 1000Genomes | |
| ENSVATH13909390 | 424 | V>I | No | 1000Genomes | |
| ENSVATH02641584 | 426 | T>M | No | 1000Genomes | |
| ENSVATH06446376 | 427 | Y>* | No | 1000Genomes | |
| ENSVATH02641586 | 428 | L>F | No | 1000Genomes | |
| tmp_4_1872097_G_C | 429 | G>R | No | 1000Genomes | |
| ENSVATH13909391 | 432 | W>L | No | 1000Genomes | |
| tmp_4_1872111_G_A,C | 433 | R>S | No | 1000Genomes | |
| ENSVATH10642669 | 435 | F>V | No | 1000Genomes | |
| tmp_4_1872127_G_A | 439 | A>T | No | 1000Genomes | |
| tmp_4_1872136_C_T | 442 | Q>* | No | 1000Genomes | |
| ENSVATH13909392 | 449 | V>L | No | 1000Genomes | |
| tmp_4_1872164_C_T | 451 | P>L | No | 1000Genomes | |
| ENSVATH06446379 | 451 | P>S | No | 1000Genomes | |
| tmp_4_1872167_C_G,T | 452 | A>G | No | 1000Genomes | |
| ENSVATH10642671 | 452 | A>T | No | 1000Genomes | |
| tmp_4_1872167_C_G,T | 452 | A>V | No | 1000Genomes | |
| ENSVATH06446381 | 457 | W>* | No | 1000Genomes | |
| tmp_4_1872187_G_A | 459 | G>R | No | 1000Genomes | |
| tmp_4_1872196_G_T,A | 462 | G>C | No | 1000Genomes | |
| tmp_4_1872196_G_T,A | 462 | G>S | No | 1000Genomes | |
| tmp_4_1872199_C_A | 463 | L>M | No | 1000Genomes | |
| ENSVATH13909393 | 465 | T>S | No | 1000Genomes | |
| tmp_4_1872212_A_G | 467 | H>R | No | 1000Genomes | |
| ENSVATH02641598 | 469 | R>C | No | 1000Genomes | |
| tmp_4_1872229_A_T | 473 | N>Y | No | 1000Genomes | |
| tmp_4_1872238_C_A | 476 | P>T | No | 1000Genomes | |
| ENSVATH08219221 | 477 | G>E | No | 1000Genomes | |
| tmp_4_1872244_G_A | 478 | A>T | No | 1000Genomes | |
| ENSVATH06446386 | 486 | W>R | No | 1000Genomes | |
| ENSVATH02641602 | 488 | G>S | No | 1000Genomes | |
| tmp_4_1872293_A_T | 494 | D>V | No | 1000Genomes | |
| ENSVATH06446387 | 497 | Q>E | No | 1000Genomes | |
| ENSVATH02641603 | 500 | E>* | No | 1000Genomes | |
| ENSVATH06446388 | 500 | E>D | No | 1000Genomes | |
| ENSVATH02641603 | 500 | E>K | No | 1000Genomes | |
| ENSVATH02641604 | 503 | V>I | No | 1000Genomes | |
| ENSVATH06446389 | 504 | A>T | No | 1000Genomes | |
| tmp_4_1872323_C_T | 504 | A>V | No | 1000Genomes | |
| ENSVATH06446390 | 505 | K>N | No | 1000Genomes | |
| ENSVATH02641605 | 506 | L>F | No | 1000Genomes | |
| ENSVATH10642674 | 513 | L>P | No | 1000Genomes | |
| tmp_4_1872353_A_T | 514 | K>I | No | 1000Genomes | |
| tmp_4_1872355_G_C | 515 | E>Q | No | 1000Genomes | |
| tmp_4_1872356_A_T | 515 | E>V | No | 1000Genomes | |
| ENSVATH02641606 | 517 | R>Q | No | 1000Genomes | |
| ENSVATH10642715 | 520 | Y>N | No | 1000Genomes | |
| ENSVATH10642716 | 521 | K>E | No | 1000Genomes | |
| tmp_4_1872377_G_A | 522 | S>N | No | 1000Genomes | |
| ENSVATH02641607 | 522 | S>R | No | 1000Genomes |
No associated diseases with Q1PEC0
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1JPL7 | PME18 | Pectinesterase/pectinesterase inhibitor 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84JX1 | PME19 | Probable pectinesterase/pectinesterase inhibitor 19 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SRX4 | PME7 | Probable pectinesterase/pectinesterase inhibitor 7 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49298 | PME6 | Probable pectinesterase/pectinesterase inhibitor 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42534 | PME2 | Pectinesterase 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q43867 | PME1 | Pectinesterase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O48711 | PME12 | Probable pectinesterase/pectinesterase inhibitor 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7Y201 | PME13 | Probable pectinesterase/pectinesterase inhibitor 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SKX2 | PME16 | Probable pectinesterase/pectinesterase inhibitor 16 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22149 | PME17 | Probable pectinesterase/pectinesterase inhibitor 17 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O80722 | PME4 | Pectinesterase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV8 | PME5 | Pectinesterase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22256 | PME20 | Probable pectinesterase/pectinesterase inhibitor 20 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GX86 | PME21 | Probable pectinesterase/pectinesterase inhibitor 21 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M9W7 | PME22 | Putative pectinesterase/pectinesterase inhibitor 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GXA1 | PME23 | Probable pectinesterase/pectinesterase inhibitor 23 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SG77 | PME24 | Putative pectinesterase/pectinesterase inhibitor 24 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9STY3 | PME33 | Probable pectinesterase/pectinesterase inhibitor 33 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LYT5 | PME35 | Probable pectinesterase/pectinesterase inhibitor 35 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84R10 | PME36 | Probable pectinesterase/pectinesterase inhibitor 36 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV6 | VGDH2 | Probable pectinesterase/pectinesterase inhibitor VGDH2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81320 | PME38 | Putative pectinesterase/pectinesterase inhibitor 38 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81415 | PME39 | Probable pectinesterase/pectinesterase inhibitor 39 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81301 | PME40 | Probable pectinesterase/pectinesterase inhibitor 40 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8RXK7 | PME41 | Probable pectinesterase/pectinesterase inhibitor 41 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O23447 | PME43 | Putative pectinesterase/pectinesterase inhibitor 43 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY7 | PME44 | Probable pectinesterase/pectinesterase inhibitor 44 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY6 | PME45 | Putative pectinesterase/pectinesterase inhibitor 45 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF78 | PME46 | Probable pectinesterase/pectinesterase inhibitor 46 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF77 | PME47 | Probable pectinesterase/pectinesterase inhibitor 47 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXD9 | PME51 | Probable pectinesterase/pectinesterase inhibitor 51 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E989 | PME54 | Probable pectinesterase/pectinesterase inhibitor 54 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E8Z8 | PME28 | Putative pectinesterase/pectinesterase inhibitor 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ21 | PME58 | Probable pectinesterase/pectinesterase inhibitor 58 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN5 | PME59 | Probable pectinesterase/pectinesterase inhibitor 59 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN4 | PME60 | Probable pectinesterase/pectinesterase inhibitor 60 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FK05 | PME61 | Probable pectinesterase/pectinesterase inhibitor 61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8L7Q7 | PME64 | Probable pectinesterase/pectinesterase inhibitor 64 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q94CB1 | PME25 | Probable pectinesterase/pectinesterase inhibitor 25 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXK7 | PME32 | Probable pectinesterase/pectinesterase inhibitor 32 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M3B0 | PME34 | Probable pectinesterase/pectinesterase inhibitor 34 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P09607 | PME2.1 | Pectinesterase 2.1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| P14280 | PME1.9 | Pectinesterase 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96576 | PME3 | Pectinesterase 3 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96575 | PME2.2 | Pectinesterase 2.2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLVKVFSFFI | LMITMVVIGV | SKEYCDDKHS | CQNFLLELKT | ASSSLSEIRR | RDLLIIVLKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SVRKIDMAMI | GVMEDTKQHE | EMENDKLCLK | EDTNLFEEMM | ESAKDRMIRS | VEELLGGEFP |
| 130 | 140 | 150 | 160 | 170 | 180 |
| YLGSYENIHT | WLSGVLTSYI | TCIDEIGDGA | YKRRVEPQLQ | DLISKAKVAL | ALFISISPRD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NTELNSVVPN | SPSWLSHVDK | KDLYLNAEAL | KKIADVVVAK | DGTGKYNTVN | AAIAAAPQHS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HKRFIIYIKT | GIYDEIVAIE | NTKPNLTLIG | DGQDSTIITG | NLSASNVRRT | FYTATFASNG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KGFIGVDMCF | RNTVGPAKGP | AVALRVSGDM | SVIYRCRVEG | YQDALYPHID | RQFYRECFIT |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GTVDFICGNA | AAVFQFCQIV | ARQPNMGQSN | FITAQSRETK | DDKSGFSIQN | CNITASSDLD |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TATVKTYLGR | PWRIFSTVAV | LQSFIGDLVD | PAGWTPWEGE | TGLSTLHYRE | YQNRGPGAVT |
| 490 | 500 | 510 | 520 | ||
| SRRVKWSGFK | VMKDPKQATE | FTVAKLLDGE | TWLKESRIPY | KSGL |