Q1JPL7
Gene name |
PME18 (ARATH4, At1g11580, T23J18.23, T23J18.24, T23J18.33) |
Protein name |
Pectinesterase/pectinesterase inhibitor 18 |
Names |
AtPMEpcrA |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G11580 |
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
Descriptions
(Annotation from UniProt)
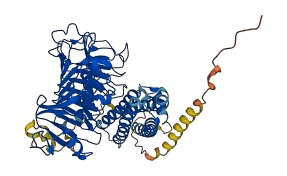
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
246-542 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for Q1JPL7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q1JPL7-F1 | Predicted | AlphaFoldDB |
33 variants for Q1JPL7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_1_3888733_T_A | 2 | S>T | No | 1000Genomes | |
| ENSVATH04558835 | 12 | P>T | No | 1000Genomes | |
| tmp_1_3888814_A_G | 29 | I>V | No | 1000Genomes | |
| ENSVATH10990769 | 34 | A>T | No | 1000Genomes | |
| tmp_1_3888867_C_G | 46 | N>K | No | 1000Genomes | |
| ENSVATH10990770 | 49 | D>G | No | 1000Genomes | |
| ENSVATH10990770 | 49 | D>V | No | 1000Genomes | |
| ENSVATH00018064 | 60 | H>Q | No | 1000Genomes | |
| tmp_1_3888925_G_C | 66 | D>H | No | 1000Genomes | |
| tmp_1_3888979_A_C | 84 | N>H | No | 1000Genomes | |
| ENSVATH13880569 | 119 | D>E | No | 1000Genomes | |
| ENSVATH00018067 | 123 | F>L | No | 1000Genomes | |
| ENSVATH10990771 | 130 | M>I | No | 1000Genomes | |
| ENSVATH04558838 | 135 | D>N | No | 1000Genomes | |
| ENSVATH01036424 | 145 | R>L | No | 1000Genomes | |
| ENSVATH00018068 | 146 | G>R | No | 1000Genomes | |
| tmp_1_3889273_T_A | 182 | S>T | No | 1000Genomes | |
| tmp_1_3889334_C_T | 202 | A>V | No | 1000Genomes | |
| tmp_1_3889345_T_A | 206 | S>T | No | 1000Genomes | |
| ENSVATH04558841 | 222 | F>Y | No | 1000Genomes | |
| tmp_1_3889433_A_T | 235 | E>V | No | 1000Genomes | |
| ENSVATH01036428 | 244 | T>I | No | 1000Genomes | |
| tmp_1_3889709_G_A | 276 | V>I | No | 1000Genomes | |
| tmp_1_3890308_A_G | 445 | T>A | No | 1000Genomes | |
| ENSVATH00018077 | 447 | S>T | No | 1000Genomes | |
| ENSVATH00018078 | 451 | A>V | No | 1000Genomes | |
| tmp_1_3890404_T_C | 477 | F>L | No | 1000Genomes | |
| ENSVATH01036431 | 484 | P>S | No | 1000Genomes | |
| ENSVATH01036431 | 484 | P>T | No | 1000Genomes | |
| tmp_1_3890454_G_T | 493 | E>D | No | 1000Genomes | |
| ENSVATH04558847 | 520 | K>N | No | 1000Genomes | |
| tmp_1_3890555_A_G | 527 | D>G | No | 1000Genomes | |
| ENSVATH10990974 | 548 | P>S | No | 1000Genomes |
No associated diseases with Q1JPL7
4 regional properties for Q1JPL7
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plant-type vacuole | A closed structure that is completely surrounded by a unit membrane, contains liquid, and retains the same shape regardless of cell cycle phase. An example of this structure is found in Arabidopsis thaliana. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
| rRNA N-glycosylase activity | Catalysis of the hydrolysis of the N-glycosylic bond at A-4324 in 28S rRNA from rat ribosomes or corresponding sites in 28S RNA from other species. |
| toxin activity | Interacting selectively with one or more biological molecules in another (target) organism, initiating pathogenesis (leading to an abnormal, generally detrimental state) in the target organism. The activity should refer to an evolved function of the active gene product, i.e. one that was selected for. Examples include the activity of botulinum toxin, and snake venom. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| defense response to fungus | Reactions triggered in response to the presence of a fungus that act to protect the cell or organism. |
| killing of cells of other organism | Any process in an organism that results in the killing of cells of another organism, including in some cases the death of the other organism. Killing here refers to the induction of death in one cell by another cell, not cell-autonomous death due to internal or other environmental conditions. |
| negative regulation of translation | Any process that stops, prevents, or reduces the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
| response to bacterium | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium. |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1PEC0 | PME42 | Probable pectinesterase/pectinesterase inhibitor 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84JX1 | PME19 | Probable pectinesterase/pectinesterase inhibitor 19 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SRX4 | PME7 | Probable pectinesterase/pectinesterase inhibitor 7 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49298 | PME6 | Probable pectinesterase/pectinesterase inhibitor 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42534 | PME2 | Pectinesterase 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q43867 | PME1 | Pectinesterase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O48711 | PME12 | Probable pectinesterase/pectinesterase inhibitor 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7Y201 | PME13 | Probable pectinesterase/pectinesterase inhibitor 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SKX2 | PME16 | Probable pectinesterase/pectinesterase inhibitor 16 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22149 | PME17 | Probable pectinesterase/pectinesterase inhibitor 17 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O80722 | PME4 | Pectinesterase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV8 | PME5 | Pectinesterase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22256 | PME20 | Probable pectinesterase/pectinesterase inhibitor 20 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GX86 | PME21 | Probable pectinesterase/pectinesterase inhibitor 21 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M9W7 | PME22 | Putative pectinesterase/pectinesterase inhibitor 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GXA1 | PME23 | Probable pectinesterase/pectinesterase inhibitor 23 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SG77 | PME24 | Putative pectinesterase/pectinesterase inhibitor 24 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9STY3 | PME33 | Probable pectinesterase/pectinesterase inhibitor 33 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LYT5 | PME35 | Probable pectinesterase/pectinesterase inhibitor 35 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84R10 | PME36 | Probable pectinesterase/pectinesterase inhibitor 36 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV6 | VGDH2 | Probable pectinesterase/pectinesterase inhibitor VGDH2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81320 | PME38 | Putative pectinesterase/pectinesterase inhibitor 38 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81415 | PME39 | Probable pectinesterase/pectinesterase inhibitor 39 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81301 | PME40 | Probable pectinesterase/pectinesterase inhibitor 40 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8RXK7 | PME41 | Probable pectinesterase/pectinesterase inhibitor 41 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O23447 | PME43 | Putative pectinesterase/pectinesterase inhibitor 43 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY7 | PME44 | Probable pectinesterase/pectinesterase inhibitor 44 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY6 | PME45 | Putative pectinesterase/pectinesterase inhibitor 45 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF78 | PME46 | Probable pectinesterase/pectinesterase inhibitor 46 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF77 | PME47 | Probable pectinesterase/pectinesterase inhibitor 47 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXD9 | PME51 | Probable pectinesterase/pectinesterase inhibitor 51 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E989 | PME54 | Probable pectinesterase/pectinesterase inhibitor 54 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E8Z8 | PME28 | Putative pectinesterase/pectinesterase inhibitor 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ21 | PME58 | Probable pectinesterase/pectinesterase inhibitor 58 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN5 | PME59 | Probable pectinesterase/pectinesterase inhibitor 59 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN4 | PME60 | Probable pectinesterase/pectinesterase inhibitor 60 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FK05 | PME61 | Probable pectinesterase/pectinesterase inhibitor 61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8L7Q7 | PME64 | Probable pectinesterase/pectinesterase inhibitor 64 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q94CB1 | PME25 | Probable pectinesterase/pectinesterase inhibitor 25 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXK7 | PME32 | Probable pectinesterase/pectinesterase inhibitor 32 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M3B0 | PME34 | Probable pectinesterase/pectinesterase inhibitor 34 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P09607 | PME2.1 | Pectinesterase 2.1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| P14280 | PME1.9 | Pectinesterase 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96576 | PME3 | Pectinesterase 3 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| Q96575 | PME2.2 | Pectinesterase 2.2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSNSNQPLLS | KPKSLKHKNL | CLVLSFVAIL | GSVAFFTAQL | ISVNTNNNDD | SLLTTSQICH |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GAHDQDSCQA | LLSEFTTLSL | SKLNRLDLLH | VFLKNSVWRL | ESTMTMVSEA | RIRSNGVRDK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AGFADCEEMM | DVSKDRMMSS | MEELRGGNYN | LESYSNVHTW | LSSVLTNYMT | CLESISDVSV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| NSKQIVKPQL | EDLVSRARVA | LAIFVSVLPA | RDDLKMIISN | RFPSWLTALD | RKLLESSPKT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LKVTANVVVA | KDGTGKFKTV | NEAVAAAPEN | SNTRYVIYVK | KGVYKETIDI | GKKKKNLMLV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GDGKDATIIT | GSLNVIDGST | TFRSATVAAN | GDGFMAQDIW | FQNTAGPAKH | QAVALRVSAD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| QTVINRCRID | AYQDTLYTHT | LRQFYRDSYI | TGTVDFIFGN | SAVVFQNCDI | VARNPGAGQK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NMLTAQGRED | QNQNTAISIQ | KCKITASSDL | APVKGSVKTF | LGRPWKLYSR | TVIMQSFIDN |
| 490 | 500 | 510 | 520 | 530 | 540 |
| HIDPAGWFPW | DGEFALSTLY | YGEYANTGPG | ADTSKRVNWK | GFKVIKDSKE | AEQFTVAKLI |
| 550 | |||||
| QGGLWLKPTG | VTFQEWL |