Q1HAQ0
Gene name |
Lpcat1 (Aytl2) |
Protein name |
Lysophosphatidylcholine acyltransferase 1 |
Names |
LPC acyltransferase 1, LPCAT-1, LysoPC acyltransferase 1, 1-acylglycerol-3-phosphate O-acyltransferase, 1-acylglycerophosphocholine O-acyltransferase, 1-alkenylglycerophosphocholine O-acyltransferase, 1-alkylglycerophosphocholine O-acetyltransferase, Acetyl-CoA:lyso-platelet-activating factor acetyltransferase, Acetyl-CoA:lyso-PAF acetyltransferase, Lyso-PAF acetyltransferase, LysoPAFAT, Acyltransferase-like 2 |
Species |
Rattus norvegicus (Rat) |
KEGG Pathway |
rno:361467 |
EC number |
2.3.1.23: Transferring groups other than amino-acyl groups |
Protein Class |
|
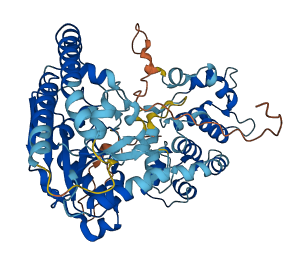
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q1HAQ0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q1HAQ0-F1 | Predicted | AlphaFoldDB |
No variants for Q1HAQ0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q1HAQ0 | |||||
No associated diseases with Q1HAQ0
No regional properties for Q1HAQ0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| No domain, repeats, and functional sites for Q1HAQ0 | |||
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.1.23 | Transferring groups other than amino-acyl groups |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| lipid droplet | An intracellular non-membrane-bounded organelle comprising a matrix of coalesced lipids surrounded by a phospholipid monolayer. May include associated proteins. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| 1-acylglycerol-3-phosphate O-acyltransferase activity | Catalysis of the reaction: acyl-CoA + 1-acyl-sn-glycerol-3-phosphate = CoA + 1,2-diacyl-sn-glycerol-3-phosphate. |
| 1-acylglycerophosphocholine O-acyltransferase activity | Catalysis of the reaction: 1-acyl-sn-glycero-3-phosphocholine + acyl-CoA = phosphatidylcholine + CoA. |
| 1-alkenylglycerophosphocholine O-acyltransferase activity | Catalysis of the reaction: 1-alkenylglycerophosphocholine + acyl-CoA = 1-alkenyl-2-acylglycerophosphocholine + CoA. |
| 1-alkylglycerophosphocholine O-acetyltransferase activity | Catalysis of the reaction: 1-alkyl-sn-glycero-3-phosphocholine + acetyl-CoA = 1-alkyl-2-acetyl-sn-glycero-3-phosphocholine + CoA. |
| 1-alkylglycerophosphocholine O-acyltransferase activity | Catalysis of the reaction: 1-alkyl-sn-glycero-3-phosphocholine + acyl-CoA = 1-alkyl-2-acyl-sn-glycero-3-phosphocholine + CoA. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| plasmalogen synthase activity | Catalysis of the reaction: acyl-CoA + 1-O-alk-1-enyl-glycero-3-phosphocholine = CoA + plasmenylcholine. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of phosphatidylcholine biosynthetic process | Any process that stops, prevents or reduces the frequency, rate or extent of phosphatidylcholine biosynthetic process. |
| phosphatidylcholine acyl-chain remodeling | Remodeling the acyl chains of phosphatidylcholine, through sequential deacylation and re-acylation reactions, to generate phosphatidylcholine containing different types of fatty acid acyl chains. |
| phosphatidylcholine biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidylcholines, any of a class of glycerophospholipids in which the phosphatidyl group is esterified to the hydroxyl group of choline. |
| phospholipid biosynthetic process | The chemical reactions and pathways resulting in the formation of a phospholipid, a lipid containing phosphoric acid as a mono- or diester. |
| positive regulation of protein catabolic process | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds. |
| protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds. |
| retina development in camera-type eye | The process whose specific outcome is the progression of the retina over time, from its formation to the mature structure. The retina is the innermost layer or coating at the back of the eyeball, which is sensitive to light and in which the optic nerve terminates. |
| surfactant homeostasis | Any process involved in the maintenance of a steady-state level of the surface-active lipoprotein mixture which coats the alveoli. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8NF37 | LPCAT1 | Lysophosphatidylcholine acyltransferase 1 | Homo sapiens (Human) | PR |
| Q6NVG1 | Lpcat4 | Lysophospholipid acyltransferase LPCAT4 | Mus musculus (Mouse) | PR |
| Q3TFD2 | Lpcat1 | Lysophosphatidylcholine acyltransferase 1 | Mus musculus (Mouse) | PR |
| Q8L7R3 | LPEAT1 | Lysophospholipid acyltransferase LPEAT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8S8S2 | LPEAT2 | Lysophospholipid acyltransferase LPEAT2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1LWG4 | lpcat1 | Lysophosphatidylcholine acyltransferase 1 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRLRGRGPRA | APSSSSGAGD | ARRLAPPGRN | PFVHELRLSA | LQKAQVAFMT | LTLFPIRLLF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AAFMMLLAWP | FALVASLGPP | DKEPEQPLAL | WRKVVDFLLK | AIMRTMWFAG | GFHRVAVKGR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QALPTEAAIL | TLAPHSSYFD | AIPVTMTMSS | IVMKAESRDI | PIWGTLIRYI | RPVFVSRSDQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DSRRKTVEEI | KRRAQSNGKW | PQIMIFPEGT | CTNRTCLITF | KPGAFIPGVP | VQPVVLRYPN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KLDTITWTWQ | GPGALKILWL | TLCQFQNQVE | IEFLPVYCPS | EEEKRNPALY | ASNVRRVMAK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ALGVSVTDYT | FEDCQLALAE | GQLRLPADTC | LLEFARLVRG | LGLKPENLEK | DLDKYSESAR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MKRGEKIRLP | EFAAYLEVPV | SDALEDMFSL | FDESGGGEID | LREYVVALSV | VCRPSQTLAT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| IQLAFKMYGS | PEDGSIDEAD | LSCILKTALG | ISELTVTDLF | QAIDQEERGR | ITFDDFCGFA |
| 490 | 500 | 510 | 520 | 530 | |
| EMYPDFAEDY | LYPDQTHSDS | CAQTPPAPTP | NGFCIDFSPE | HSDFGRKNSC | KKVD |