Q18486
Gene name |
coq-8 (C35D10.4) |
Protein name |
Atypical kinase coq-8, mitochondrial |
Names |
EC 2.7.-.- |
Species |
Caenorhabditis elegans |
KEGG Pathway |
cel:CELE_C35D10.4 |
EC number |
2.7.-.-: Transferring phosphorus-containing groups |
Protein Class |
|
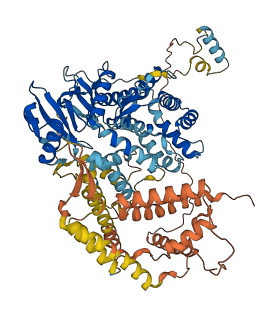
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
394-648 (Activator of bc1 complex, ABC1, kinases, also called aarF domain containing kinase 3) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q18486
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q18486-F1 | Predicted | AlphaFoldDB |
No variants for Q18486
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q18486 | |||||
No associated diseases with Q18486
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.-.- | Transferring phosphorus-containing groups |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| transferase activity | Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| ubiquinone biosynthetic process | The chemical reactions and pathways resulting in the formation of ubiquinone, a lipid-soluble electron-transporting coenzyme. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P27697 | COQ8 | Atypical kinase COQ8, mitochondrial | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | SS |
| Q29RI0 | COQ8A | Atypical kinase COQ8A, mitochondrial | Bos taurus (Bovine) | SS |
| Q96D53 | COQ8B | Atypical kinase COQ8B, mitochondrial | Homo sapiens (Human) | SS |
| Q8NI60 | COQ8A | Atypical kinase COQ8A, mitochondrial | Homo sapiens (Human) | EV |
| Q60936 | Coq8a | Atypical kinase COQ8A, mitochondrial | Mus musculus (Mouse) | SS |
| Q566J8 | Coq8b | Atypical kinase COQ8B, mitochondrial | Mus musculus (Mouse) | SS |
| Q5BJQ0 | Coq8a | Atypical kinase COQ8A, mitochondrial | Rattus norvegicus (Rat) | SS |
| Q6AY19 | Coq8b | Atypical kinase COQ8B, mitochondrial | Rattus norvegicus (Rat) | SS |
| Q9SBB2 | ABC1 | Protein ABC transporter 1, mitochondrial | Arabidopsis thaliana (Mouse-ear cress) | SS |
| A3QJU3 | coq8b | Atypical kinase COQ8B, mitochondrial | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| Q5RGU1 | coq8a | Atypical kinase COQ8A, mitochondrial | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAKRPPAST | LKWCQQEMGP | ICEGIGMVVR | SQIGYDGRRI | EKQIRRKALQ | TVVMGGQDVD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PLKEPNKTNA | PLLSPTLPST | NEALQRARVV | AAGIETFAKL | MSQGTYPGSA | GFTVSETGER |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KYMKQTSPVQ | SNIVANLMAT | ADKLSKGVIS | LAPFPLPTIT | KLGTWVIANP | TAQLPIPPQL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KSAIPAIGQI | ETLIQSILGV | QGPVSSVSRA | PRKLKLEGVT | DQVVLEPETI | HATVDVEPID |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KTLTKEEAEF | LIKAARSVDD | DNAETAIFGT | QKAKPKVYRP | ELPKNFEVNL | EMGNVHTLTR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SNESSVPATR | IGRLATFGQL | AFGLLGGATA | EVTRRTFGIG | KRLQEEGIPK | NPFLSEANAD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RIVATLCRVR | GAALKLGQML | SIQDSSTVPP | ALLQIFERVR | QSADFMPIKQ | VHRQMKDAFG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DDWREKFEHF | DDKPFACASI | GQVHKAVLKD | GRNVAVKVQY | PGVAEGIDSD | IDNLVSVLSV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GGIFPKGMFL | DAFVGVARRE | LKQECDYERE | ARAMKKFREL | IADWQDVYVP | EVIDELSSSR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VLTTELVYGK | PVDACVEEPQ | VVRDYIAGKF | IELCLKEIFL | WRFMQTDPNW | SNFFLGKHPK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TGEPRLVLLD | FGASRAYGKK | FVDIYMNIIK | SAYDGDKKKI | IEYSREIGFL | TGYETSVMED |
| 670 | 680 | 690 | 700 | 710 | 720 |
| AHVESVMIMG | ETLASNHPYN | FANQDVTMRI | QKLIPVMLEH | RLTSPPEEIY | SLHRKLSGCY |
| 730 | 740 | 750 | |||
| LLAAKLKATV | SCGGLFHEIH | ENYVFGEDGI | DINID |