Q17941
Gene name |
akt-1 |
Protein name |
Serine/threonine-protein kinase akt-1 |
Names |
Protein kinase B akt-1, PKB akt-1 |
Species |
Caenorhabditis elegans |
KEGG Pathway |
cel:CELE_C12D8.10 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
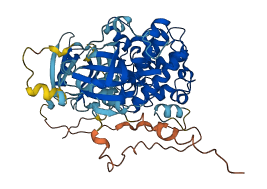
Descriptions
(Annotation based on sequence homology with P31749)
The protein kinase Akt is one of the primary effectors of growth factor signaling in the cell. Akt responds specifically to the lipid second messengers phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3] and phosphatidylinositol-3,4-bisphosphate [PI(3,4)P2] via its autoinhibitory domain (PH domain). Recruitment of Akt to PI(3,4,5)P3 in the plasma membrane promotes its phosphorylation by phosphoinositide-dependent kinase 1 (PDK1) in its activation loop (T308). Phosphorylation of S473 within AGC kinase C-terminal domain activates Akt through the formation of an electrostatic interaction with a conserved basic residue (R144) in the PH-kinase domain linker, thereby relieving PH domain- mediated autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
333-356 (Activation loop from InterPro)
Target domain |
193-450 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for Q17941
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q17941-F1 | Predicted | AlphaFoldDB |
No variants for Q17941
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q17941 | |||||
No associated diseases with Q17941
7 regional properties for Q17941
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 193 - 450 | IPR000719 |
| domain | AGC-kinase, C-terminal | 451 - 528 | IPR000961 |
| domain | Pleckstrin homology domain | 15 - 120 | IPR001849 |
| active_site | Serine/threonine-protein kinase, active site | 312 - 324 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 199 - 222 | IPR017441 |
| domain | Protein kinase, C-terminal | 471 - 518 | IPR017892 |
| domain | Protein Kinase B, pleckstrin homology domain | 14 - 121 | IPR039026 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| axon | The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter. |
| cytoplasmic side of plasma membrane | The leaflet the plasma membrane that faces the cytoplasm and any proteins embedded or anchored in it or attached to its surface. |
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| protein kinase complex | A protein complex which is capable of protein kinase activity. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| metal ion binding | Binding to a metal ion. |
| phosphatidylinositol-3,4,5-trisphosphate binding | Binding to phosphatidylinositol-3,4,5-trisphosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 3', 4' and 5' positions. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| chemosensory behavior | Behavior that is dependent upon the sensation of chemicals. |
| chemotaxis | The directed movement of a motile cell or organism, or the directed growth of a cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| determination of adult lifespan | The pathways that regulate the duration of the adult phase of the life-cycle of an animal. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| insulin receptor signaling pathway | The series of molecular signals generated as a consequence of the insulin receptor binding to insulin. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | The series of molecular signals in which an intracellular signal is conveyed to trigger the apoptotic death of a cell. The pathway is induced by the cell cycle regulator phosphoprotein p53, or an equivalent protein, in response to the detection of DNA damage, and ends when the execution phase of apoptosis is triggered. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| peptidyl-threonine phosphorylation | The phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of synaptic assembly at neuromuscular junction | Any process that modulates the frequency, rate or extent of synaptic assembly at neuromuscular junctions. |
| response to salt | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a salt stimulus. |
17 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q01314 | AKT1 | RAC-alpha serine/threonine-protein kinase | Bos taurus (Bovine) | SS |
| Q8INB9 | Akt | RAC serine/threonine-protein kinase | Drosophila melanogaster (Fruit fly) | SS |
| P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| P31751 | AKT2 | RAC-beta serine/threonine-protein kinase | Homo sapiens (Human) | EV SS |
| Q9Y243 | AKT3 | RAC-gamma serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| P31750 | Akt1 | RAC-alpha serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q60823 | Akt2 | RAC-beta serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| Q9WUA6 | Akt3 | RAC-gamma serine/threonine-protein kinase | Mus musculus (Mouse) | PR |
| P47196 | Akt1 | RAC-alpha serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| P47197 | Akt2 | RAC-beta serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| Q63484 | Akt3 | RAC-gamma serine/threonine-protein kinase | Rattus norvegicus (Rat) | PR |
| Q9XTG7 | akt-2 | Serine/threonine-protein kinase akt-2 | Caenorhabditis elegans | SS |
| Q19266 | pkc-3 | Protein kinase C-like 3 | Caenorhabditis elegans | SS |
| Q9Y1J3 | pdk-1 | 3-phosphoinositide-dependent protein kinase 1 | Caenorhabditis elegans | SS |
| P34722 | tpa-1 | Protein kinase C-like 1 | Caenorhabditis elegans | PR |
| P90980 | pkc-2 | Protein kinase C-like 2 | Caenorhabditis elegans | SS |
| Q2L6W9 | sax-1 | Serine/threonine-protein kinase sax-1 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSMTSLSTKS | RRQEDVVIEG | WLHKKGEHIR | NWRPRYFMIF | NDGALLGFRA | KPKEGQPFPE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PLNDFMIKDA | ATMLFEKPRP | NMFMVRCLQW | TTVIERTFYA | ESAEVRQRWI | HAIESISKKY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KGTNANPQEE | LMETNQQPKI | DEDSEFAGAA | HAIMGQPSSG | HGDNCSIDFR | ASMISIADTS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EAAKRDKITM | EDFDFLKVLG | KGTFGKVILC | KEKRTQKLYA | IKILKKDVII | AREEVAHTLT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ENRVLQRCKH | PFLTELKYSF | QEQHYLCFVM | QFANGGELFT | HVRKCGTFSE | PRARFYGAEI |
| 310 | 320 | 330 | 340 | 350 | 360 |
| VLALGYLHRC | DIVYRDMKLE | NLLLDKDGHI | KIADFGLCKE | EISFGDKTST | FCGTPEYLAP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EVLDDHDYGR | CVDWWGVGVV | MYEMMCGRLP | FYSKDHNKLF | ELIMAGDLRF | PSKLSQEART |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LLTGLLVKDP | TQRLGGGPED | ALEICRADFF | RTVDWEATYR | KEIEPPYKPN | VQSETDTSYF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| DNEFTSQPVQ | LTPPSRSGAL | ATVDEQEEMQ | SNFTQFSFHN | VMGSINRIHE | ASEDNEDYDM |
| G |