Q16763
Gene name |
UBE2S (E2EPF, OK/SW-cl.73) |
Protein name |
Ubiquitin-conjugating enzyme E2 S |
Names |
EC 2.3.2.23 , E2 ubiquitin-conjugating enzyme S , E2-EPF , Ubiquitin carrier protein S , Ubiquitin-conjugating enzyme E2-24 kDa , Ubiquitin-conjugating enzyme E2-EPF5 , Ubiquitin-protein ligase S |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:27338 |
EC number |
2.3.2.23: Aminoacyltransferases |
Protein Class |
UBIQUITIN-CONJUGATING ENZYME E2 (PTHR24067) |
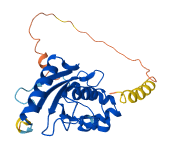
Descriptions
Ubiquitin conjugating enzyme E2 S (UBE2S) is a protein that plays a key role in accepting ubiquitin from the E1 complex and catalyzing its covalent attachment to other proteins. Dimerization regulates UBE2S by preventing autoubiquitination, thus stabilizing the protein and allowing it to inhibit premature ubiquitin chain elongation during the cell cycle. The autoinhibition is mediated by blocking the ubiquitin-binding sites in UBC domain, which are essential for its E2 activity. The C-helix of UBE2S engages the APC/C for chain elongation, blocking the promotion of UBE2S autoinhibition through dimerization achieved by C-helix.
Autoinhibitory domains (AIDs)
Target domain |
1-156 (UBC domain) |
Relief mechanism |
PTM |
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
No accessory elements
References
- Holliday MJ et al. (2019) "Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation", Nature communications, 10, 3070
- Liess AKL et al. (2020) "Dimerization regulates the human APC/C-associated ubiquitin-conjugating enzyme UBE2S", Science signaling, 13,
- Bremm A (2020) "Hug and hold tight: Dimerization controls the turnover of the ubiquitin-conjugating enzyme UBE2S", Science signaling, 13,
Autoinhibited structure

Activated structure

10 structures for Q16763
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1ZDN | X-ray | 193 A | A/B | 1-156 | PDB |
| 5BNB | X-ray | 249 A | A/B/C/D | 1-156 | PDB |
| 5L9T | EM | 640 A | T | 1-222 | PDB |
| 6QH3 | X-ray | 290 A | A/B | 1-156 | PDB |
| 6QHK | X-ray | 196 A | A/B | 1-156 | PDB |
| 6S96 | X-ray | 218 A | A/B | 1-156 | PDB |
| 6S98 | X-ray | 155 A | A/B | 1-156 | PDB |
| 7AHF | X-ray | 215 A | A/B | 1-156 | PDB |
| 8TAU | EM | 350 A | E | 205-222 | PDB |
| AF-Q16763-F1 | Predicted | AlphaFoldDB |
224 variants for Q16763
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs756893985 | 4 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs756893985 | 4 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs756893985 | 4 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs2090099963 | 5 | V>M | No |
TOPMed gnomAD |
|
| rs1452269907 | 9 | P>A | No |
TOPMed gnomAD |
|
| rs552136391 | 9 | P>L | No | gnomAD | |
| rs552136391 | 9 | P>R | No | gnomAD | |
| rs1452269907 | 9 | P>S | No |
TOPMed gnomAD |
|
| rs1453830867 | 10 | P>L | No | gnomAD | |
| COSM6152223 | 13 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1195786947 | 13 | I>T | No | gnomAD | |
|
rs374069290 COSM4921770 |
14 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP NCI-TCGA TOPMed |
| rs150992671 | 14 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374069290 | 14 | R>S | No |
ESP TOPMed |
|
| rs531211876 | 15 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs921581783 | 18 | K>E | No | TOPMed | |
| rs1568442369 | 19 | E>V | No | Ensembl | |
| rs767514205 | 21 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs762040182 | 21 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2123317306 | 22 | T>I | No | Ensembl | |
| rs370647044 | 23 | L>V | No | ESP | |
| rs1308392428 | 24 | T>I | No |
TOPMed gnomAD |
|
| rs1030465280 | 25 | A>T | No |
TOPMed gnomAD |
|
| rs1234661620 | 26 | D>E | No | gnomAD | |
| rs1288541692 | 27 | P>L | No |
TOPMed gnomAD |
|
| rs2090099388 | 29 | D>N | No | TOPMed | |
| rs762797582 | 34 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs2090099325 | 35 | P>A | No | Ensembl | |
| rs2090099325 | 35 | P>S | No | Ensembl | |
| rs1286125979 | 36 | N>I | No |
TOPMed gnomAD |
|
| rs139337996 | 36 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs376302634 | 37 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs1299385628 | 37 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs745825218 | 38 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs770350118 | 39 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs780705474 | 39 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs770350118 | 39 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs997693683 | 40 | L>H | No | Ensembl | |
| rs1251231304 | 40 | L>V | No | TOPMed | |
| rs1439506585 | 41 | T>S | No |
TOPMed gnomAD |
|
| rs2090099057 | 45 | V>I | No | TOPMed | |
| rs187433678 | 46 | T>A | No | 1000Genomes | |
| rs2090099014 | 48 | E>K | No |
TOPMed gnomAD |
|
| rs2090083578 | 52 | G>E | No | Ensembl | |
| rs746549935 | 52 | G>W | No |
ExAC gnomAD |
|
| rs1286505172 | 59 | L>V | No | gnomAD | |
| rs2090083485 | 60 | F>S | No | gnomAD | |
| rs747823368 | 61 | R>C | No |
ExAC gnomAD |
|
| rs780505915 | 61 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1402132763 | 62 | M>I | No | TOPMed | |
| rs2090083430 | 62 | M>L | No | TOPMed | |
| rs868786422 | 63 | K>E | No | Ensembl | |
| rs1415474642 | 63 | K>T | No |
TOPMed gnomAD |
|
| rs1265652780 | 71 | P>L | No | gnomAD | |
| rs2090083301 | 72 | A>D | No | Ensembl | |
| rs1393368757 | 73 | S>F | No |
TOPMed gnomAD |
|
| rs2090083229 | 76 | K>R | No | TOPMed | |
| rs2090083194 | 78 | Y>F | No | TOPMed | |
| rs1308036650 | 80 | L>V | No |
TOPMed gnomAD |
|
| rs781611407 | 86 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs758572803 | 88 | V>M | No |
ExAC gnomAD |
|
| rs1049501918 | 89 | G>V | No | Ensembl | |
|
rs765011011 COSM3797556 |
90 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs765011011 | 90 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2090082886 | 91 | N>H | No | gnomAD | |
| rs540315809 | 91 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs766266228 | 92 | G>D | No |
ExAC gnomAD |
|
|
COSM5572035 rs1350509514 |
96 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1439694374 | 97 | N>S | No |
TOPMed gnomAD |
|
| rs747733579 | 98 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2090082726 | 100 | K>E | No | gnomAD | |
| rs774217206 | 100 | K>R | No |
ExAC gnomAD |
|
| rs770184703 | 102 | D>E | No |
ExAC gnomAD |
|
| TCGA novel | 102 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2090082684 | 104 | T>M | No | TOPMed | |
| TCGA novel | 105 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781704539 | 108 | G>D | No |
ExAC gnomAD |
|
| rs1400054219 | 110 | R>* | No |
TOPMed gnomAD |
|
| rs1382629460 | 111 | H>Q | No |
TOPMed gnomAD |
|
| rs1330328877 | 112 | V>A | No |
TOPMed gnomAD |
|
| rs771304310 | 112 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2090060796 | 115 | T>I | No | Ensembl | |
| rs1600318089 | 115 | T>P | No | Ensembl | |
| rs573318166 | 117 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1600318065 | 122 | H>P | No | Ensembl | |
| rs2090060677 | 123 | P>A | No | TOPMed | |
| rs1600318061 | 124 | N>T | No | Ensembl | |
| rs746917958 | 125 | P>A | No |
ExAC gnomAD |
|
| rs370428278 | 126 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs1395975605 | 129 | L>F | No | gnomAD | |
| rs764747972 | 130 | N>S | No |
ExAC gnomAD |
|
| rs753488671 | 131 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs753488671 | 131 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1471768723 | 133 | A>T | No | gnomAD | |
| rs1369706768 | 133 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1600317988 | 134 | G>A | No | Ensembl | |
| rs2090060274 | 134 | G>S | No | TOPMed | |
| rs1235935851 | 135 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs762293175 COSM4127907 |
135 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2090060157 | 136 | L>P | No |
TOPMed gnomAD |
|
| rs763404918 | 138 | L>F | No |
ExAC gnomAD |
|
| rs1569048131 | 140 | N>D | No | Ensembl | |
| rs775594881 | 141 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs775594881 | 141 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs2090060028 | 142 | E>G | No | TOPMed | |
| rs759766193 | 142 | E>K | No |
ExAC gnomAD |
|
| rs759766193 | 142 | E>Q | No |
ExAC gnomAD |
|
| rs374167114 | 145 | A>T | No |
ESP gnomAD |
|
| rs1357491467 | 145 | A>V | No |
TOPMed gnomAD |
|
| rs2090059924 | 146 | A>T | No | TOPMed | |
| rs771995980 | 147 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs771995980 | 147 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs777458627 | 147 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs748048525 | 148 | A>S | No | ExAC | |
| rs778323528 | 149 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs754546129 | 149 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs757533997 | 155 | H>Y | No |
ExAC gnomAD |
|
| rs1292690585 | 156 | G>R | No | gnomAD | |
| rs753261540 | 157 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs765404886 | 157 | G>D | No |
ExAC gnomAD |
|
| rs753261540 | 157 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs771143319 | 158 | A>D | No |
ExAC gnomAD |
|
| rs556845857 | 158 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 158 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1312646281 | 159 | G>A | No |
TOPMed gnomAD |
|
| rs773001900 | 159 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1312646281 | 159 | G>V | No |
TOPMed gnomAD |
|
| rs200933021 | 160 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs768128796 | 161 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs768128796 | 161 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs778875694 | 161 | P>S | No | ExAC | |
| rs1207684429 | 162 | S>G | No | TOPMed | |
| rs748838927 | 162 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs750061593 | 163 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs750061593 | 163 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs750061593 | 163 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs778132592 | 163 | G>V | No |
ExAC gnomAD |
|
| rs758756486 | 165 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs765896389 | 166 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1034316727 | 167 | A>S | No | Ensembl | |
| rs766382400 | 168 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs753918789 | 168 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs766382400 | 168 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs767327662 | 169 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs534399153 | 169 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs534399153 | 169 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767327662 | 169 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs774238958 | 170 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1217946445 | 170 | A>V | No | gnomAD | |
| rs1338267762 | 172 | A>T | No |
TOPMed gnomAD |
|
| rs768543897 | 173 | S>G | No |
ExAC gnomAD |
|
| rs1306615876 | 173 | S>N | No | TOPMed | |
| rs768543897 | 173 | S>R | No |
ExAC gnomAD |
|
| rs749248836 | 174 | G>D | No |
ExAC gnomAD |
|
| rs749248836 | 174 | G>V | No |
ExAC gnomAD |
|
| rs2090058877 | 175 | T>I | No | Ensembl | |
|
RCV000950018 rs61748180 |
176 | E>A | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs745530942 | 177 | A>D | No |
ExAC TOPMed gnomAD |
|
| rs1415815212 | 177 | A>T | No |
TOPMed gnomAD |
|
| rs745530942 | 177 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs780766273 | 179 | S>C | No |
ExAC gnomAD |
|
| rs780766273 | 179 | S>F | No |
ExAC gnomAD |
|
| rs377751817 | 181 | D>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779440236 | 182 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs755484248 | 183 | G>R | No |
ExAC gnomAD |
|
| rs2090058568 | 184 | A>T | No |
TOPMed gnomAD |
|
| rs1373745648 | 184 | A>V | No | gnomAD | |
| rs1212921567 | 185 | P>A | No |
TOPMed gnomAD |
|
| rs766472486 | 186 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs762051776 | 187 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs2090058388 | 187 | G>C | No | TOPMed | |
| rs762051776 | 187 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs2090058388 | 187 | G>S | No | TOPMed | |
| rs762051776 | 187 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs201696148 | 188 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201696148 | 188 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs953767072 | 188 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs762811480 | 189 | G>E | No |
ExAC gnomAD |
|
| rs1281856284 | 189 | G>R | No | gnomAD | |
|
rs897520466 COSM3539451 |
190 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs745422668 | 192 | E>K | No |
ExAC gnomAD |
|
| COSM6152224 | 193 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1318615162 | 193 | G>V | No |
TOPMed gnomAD |
|
| rs2090058018 | 195 | M>I | No |
TOPMed gnomAD |
|
| rs746669366 | 195 | M>T | No |
ExAC gnomAD |
|
| rs770655715 | 195 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1437182817 | 196 | A>T | No | TOPMed | |
| rs2090057914 | 199 | H>R | No | TOPMed | |
| rs994456561 | 199 | H>Y | No |
TOPMed gnomAD |
|
| rs1005962215 | 201 | G>V | No | Ensembl | |
| rs749798764 | 202 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs780473933 | 203 | R>C | No |
ExAC gnomAD |
|
| rs756187319 | 203 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs757414025 | 204 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs767659246 | 204 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs767659246 | 204 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs767659246 | 204 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs1379223748 | 206 | K>N | No | gnomAD | |
| rs2090057706 | 206 | K>Q | No | Ensembl | |
| rs1444111571 | 208 | A>T | No |
TOPMed gnomAD |
|
| rs373412596 | 208 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2090057565 | 209 | A>T | No |
TOPMed gnomAD |
|
| rs762799159 | 209 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs752661607 | 211 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1487657978 | 211 | K>R | No |
TOPMed gnomAD |
|
| rs1407201235 | 213 | T>A | No |
TOPMed gnomAD |
|
| rs376584188 | 213 | T>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs376584188 | 213 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1437987468 | 214 | D>E | No |
TOPMed gnomAD |
|
| rs1600317397 | 215 | K>E | No | Ensembl | |
| rs770567371 | 216 | K>Q | No |
ExAC gnomAD |
|
| rs528548172 | 217 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs769119945 | 217 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs528548172 | 217 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs749653893 | 218 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs780497000 | 218 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs559435005 | 220 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs781182726 | 220 | R>W | No |
ExAC gnomAD |
|
| rs953632326 | 221 | R>Q | No | TOPMed | |
| rs1311954672 | 221 | R>W | No | gnomAD | |
| rs2090057114 | 222 | L>P | No | Ensembl | |
| rs1392371922 | 222 | L>V | No | gnomAD | |
| rs2123306894 | 223 | L>E | No | Ensembl | |
| rs1294721857 | 223 | L>L | No | Ensembl |
No associated diseases with Q16763
14 regional properties for Q16763
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 518 - 790 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 519 - 786 | IPR001245 |
| domain | Immunoglobulin subtype 2 | 55 - 124 | IPR003598-1 |
| domain | Immunoglobulin subtype 2 | 151 - 210 | IPR003598-2 |
| domain | Immunoglobulin subtype | 49 - 136 | IPR003599-1 |
| domain | Immunoglobulin subtype | 145 - 222 | IPR003599-2 |
| domain | Fibronectin type III | 225 - 320 | IPR003961-1 |
| domain | Fibronectin type III | 323 - 416 | IPR003961-2 |
| domain | Immunoglobulin-like domain | 36 - 128 | IPR007110-1 |
| domain | Immunoglobulin-like domain | 139 - 220 | IPR007110-2 |
| active_site | Tyrosine-protein kinase, active site | 651 - 663 | IPR008266 |
| domain | Immunoglobulin I-set | 48 - 124 | IPR013098 |
| binding_site | Protein kinase, ATP binding site | 524 - 550 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 518 - 786 | IPR020635 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.23 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24067 | UBIQUITIN-CONJUGATING ENZYME E2 |
| PANTHER Subfamily | PTHR24067:SF154 | UBIQUITIN-CONJUGATING ENZYME E2 G2 |
| PANTHER Protein Class |
ubiquitin-protein ligase
protein modifying enzyme |
|
| PANTHER Pathway Category |
Parkinson disease Ubc7 Ubiquitin proteasome pathway E2 |
|
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| anaphase-promoting complex | A ubiquitin ligase complex that degrades mitotic cyclins and anaphase inhibitory protein, thereby triggering sister chromatid separation and exit from mitosis. Substrate recognition by APC occurs through degradation signals, the most common of which is termed the Dbox degradation motif, originally discovered in cyclin B. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| anaphase-promoting complex binding | Binding to an anaphase-promoting complex. A ubiquitin ligase complex that degrades mitotic cyclins and anaphase inhibitory protein, thereby triggering sister chromatid separation and exit from mitosis. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ubiquitin conjugating enzyme activity | Isoenergetic transfer of ubiquitin from one protein to another via the reaction X-ubiquitin + Y -> Y-ubiquitin + X, where both the X-ubiquitin and Y-ubiquitin linkages are thioester bonds between the C-terminal glycine of ubiquitin and a sulfhydryl side group of a cysteine residue. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| anaphase-promoting complex-dependent catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, with ubiquitin-protein ligation catalyzed by the anaphase-promoting complex, and mediated by the proteasome. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| exit from mitosis | The cell cycle transition where a cell leaves M phase and enters a new G1 phase. M phase is the part of the mitotic cell cycle during which mitosis and cytokinesis take place. |
| free ubiquitin chain polymerization | The process of creating free ubiquitin chains, compounds composed of a large number of ubiquitin monomers. These chains are not conjugated to a protein. |
| positive regulation of ubiquitin protein ligase activity | Any process that activates or increases the frequency, rate or extent of ubiquitin protein ligase activity. |
| protein K11-linked ubiquitination | A protein ubiquitination process in which ubiquitin monomers are attached to a protein, and then ubiquitin polymers are formed by linkages between lysine residues at position 11 of the ubiquitin monomers. K11-linked polyubiquitination targets the substrate protein for degradation. The anaphase-promoting complex promotes the degradation of mitotic regulators by assembling K11-linked polyubiquitin chains. |
| protein K27-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 27 of the ubiquitin monomers, is added to a protein. |
| protein K29-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 29 of the ubiquitin monomers, is added to a protein. K29-linked ubiquitination targets the substrate protein for degradation. |
| protein K6-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 6 of the ubiquitin monomers, is added to a protein. K6-linked ubiquitination is involved in DNA repair. |
| protein K63-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 63 of the ubiquitin monomers, is added to a protein. K63-linked ubiquitination does not target the substrate protein for degradation, but is involved in several pathways, notably as a signal to promote error-free DNA postreplication repair. |
| protein modification process | The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification). |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
17 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q1RML1 | UBE2S | Ubiquitin-conjugating enzyme E2 S | Bos taurus (Bovine) | SS |
| Q9VX25 | CG8188 | Ubiquitin-conjugating enzyme E2 S | Drosophila melanogaster (Fruit fly) | SS |
| Q9NPD8 | UBE2T | Ubiquitin-conjugating enzyme E2 T | Homo sapiens (Human) | PR |
| Q96LR5 | UBE2E2 | Ubiquitin-conjugating enzyme E2 E2 | Homo sapiens (Human) | PR |
| Q969T4 | UBE2E3 | Ubiquitin-conjugating enzyme E2 E3 | Homo sapiens (Human) | PR |
| Q9Y385 | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | Homo sapiens (Human) | PR |
| Q9H8T0 | AKTIP | AKT-interacting protein | Homo sapiens (Human) | PR |
| P61086 | UBE2K | Ubiquitin-conjugating enzyme E2 K | Homo sapiens (Human) | PR |
| O00762 | UBE2C | Ubiquitin-conjugating enzyme E2 C | Homo sapiens (Human) | PR |
| O14933 | UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | Homo sapiens (Human) | PR |
| Q7Z7E8 | UBE2Q1 | Ubiquitin-conjugating enzyme E2 Q1 | Homo sapiens (Human) | PR |
| A1L167 | UBE2QL1 | Ubiquitin-conjugating enzyme E2Q-like protein 1 | Homo sapiens (Human) | PR |
| Q921J4 | Ube2s | Ubiquitin-conjugating enzyme E2 S | Mus musculus (Mouse) | SS |
| B5DFI8 | Ube2s | Ubiquitin-conjugating enzyme E2 S | Rattus norvegicus (Rat) | SS |
| Q9FF66 | UBC22 | Ubiquitin-conjugating enzyme E2 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q28F89 | ube2s | Ubiquitin-conjugating enzyme E2 S | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q4V908 | ube2s | Ubiquitin-conjugating enzyme E2 S | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNSNVENLPP | HIIRLVYKEV | TTLTADPPDG | IKVFPNEEDL | TDLQVTIEGP | EGTPYAGGLF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RMKLLLGKDF | PASPPKGYFL | TKIFHPNVGA | NGEICVNVLK | RDWTAELGIR | HVLLTIKCLL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IHPNPESALN | EEAGRLLLEN | YEEYAARARL | LTEIHGGAGG | PSGRAEAGRA | LASGTEASST |
| 190 | 200 | 210 | 220 | ||
| DPGAPGGPGG | AEGPMAKKHA | GERDKKLAAK | KKTDKKRALR | RL |