Q16695
Gene name |
H3-4 |
Protein name |
Histone H3.1t |
Names |
H3/t, H3t, H3/g, Histone H3.4 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8290 |
EC number |
|
Protein Class |
|
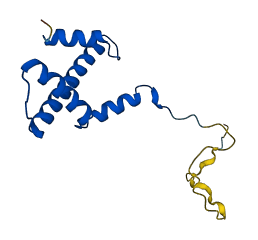
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

11 structures for Q16695
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2V1D | X-ray | 310 A | C | 2-22 | PDB |
| 2YBP | X-ray | 202 A | C/D | 31-42 | PDB |
| 2YBS | X-ray | 232 A | C/D | 31-42 | PDB |
| 3A6N | X-ray | 270 A | A/E | 1-136 | PDB |
| 3T6R | X-ray | 195 A | D | 2-8 | PDB |
| 4V2V | X-ray | 200 A | C/D | 26-30 | PDB |
| 4V2W | X-ray | 181 A | C | 17-36 | PDB |
| 6OIE | X-ray | 208 A | C/D | 2-20 | PDB |
| 6WAT | X-ray | 180 A | A/B/C/D/E/F/G/H/I/J/K/L/M/N/O/P/Q/R/S/T/V/Y/Z/a/b/c/d/e/f/g | 22-30 | PDB |
| 6WAU | X-ray | 175 A | G/H/I/J/K/L | 22-33 | PDB |
| AF-Q16695-F1 | Predicted | AlphaFoldDB |
233 variants for Q16695
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA345144764 rs774804205 |
2 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441481 rs759975640 |
2 | A>P | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441480 rs759975640 |
2 | A>S | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441479 rs774804205 |
2 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441477 rs150247684 |
3 | R>* | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC TOPMed ClinGen NCI-TCGA |
|
CA345144757 rs150247684 |
3 | R>G | No |
1000Genomes ESP ExAC TOPMed ClinGen |
|
|
rs773567566 CA345144756 |
3 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs773567566 CA1441476 |
3 | R>P | No |
ExAC gnomAD ClinGen |
|
|
rs773567566 CA345144755 COSM905191 |
3 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs748123791 CA1441473 |
4 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345144735 rs374675557 |
4 | T>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs748123791 CA345144743 |
4 | T>P | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441472 rs374675557 |
4 | T>S | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA345144721 rs1422648667 |
5 | K>E | No |
gnomAD ClinGen |
|
|
rs1387025845 CA345144712 |
5 | K>R | No |
ClinGen gnomAD |
|
|
CA1441469 rs779875414 |
6 | Q>L | No |
ExAC ClinGen |
|
|
rs567472251 CA345144667 |
7 | T>I | No |
gnomAD ClinGen |
|
|
rs567472251 CA38748203 |
7 | T>S | No |
gnomAD ClinGen |
|
|
CA345144655 rs375918041 |
8 | A>E | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441468 rs114860054 |
8 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1441467 rs375918041 |
8 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441465 rs147766330 |
9 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373549345 CA1441464 |
9 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA1441462 rs373549345 |
9 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373549345 CA1441463 |
9 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345144583 rs752078278 CA345144586 |
11 | S>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1208768158 CA345144597 |
11 | S>A | No |
TOPMed ClinGen |
|
|
rs752078278 CA1441460 |
11 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA38748173 rs1041442759 |
12 | T>A | No |
Ensembl ClinGen |
|
|
rs1410959088 CA345144570 |
12 | T>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA1441456 rs765314078 |
13 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345144550 rs765314078 |
13 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1397772335 CA345144558 |
13 | G>R | No |
TOPMed gnomAD ClinGen |
|
|
rs1397772335 CA345144561 |
13 | G>S | No |
TOPMed gnomAD ClinGen |
|
|
CA1441454 rs567618899 |
14 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1441452 rs369770019 |
15 | K>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs547680085 CA1441453 |
15 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
CA345144487 rs1324149897 |
16 | A>T | No |
gnomAD ClinGen |
|
|
rs571515258 CA1441451 |
16 | A>V | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs151071964 CA1441449 |
17 | P>L | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
rs151071964 CA345144459 COSM3385874 |
17 | P>Q | pancreas [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs551314787 CA1441448 |
18 | R>C | No |
1000Genomes ExAC gnomAD ClinGen |
|
|
rs756810829 CA1441447 |
18 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD ClinGen NCI-TCGA |
|
CA345144424 rs756810829 |
18 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756810829 CA38748135 |
18 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs369767840 CA1441446 |
19 | K>R | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA1441445 rs777184854 |
20 | Q>* | No |
ExAC ClinGen |
|
|
rs375264894 CA38748116 |
22 | A>T | No |
ESP TOPMed gnomAD ClinGen |
|
|
CA1441444 rs755642778 |
22 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA38748113 rs910077099 |
23 | T>I | No |
TOPMed gnomAD ClinGen |
|
|
CA345144332 rs910077099 |
23 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1574109655 CA345144308 |
24 | K>R | No |
ClinGen Ensembl |
|
| TCGA novel | 24 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1441442 rs372401201 |
25 | V>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345144296 rs372401201 |
25 | V>M | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
rs758842988 CA1441441 |
26 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs750781461 CA1441440 |
27 | R>C | No |
ExAC TOPMed gnomAD ClinGen |
|
|
COSM905190 CA1441439 rs765519082 |
27 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ExAC TOPMed gnomAD ClinGen cosmic curated NCI-TCGA |
|
CA345144232 rs765519082 |
27 | R>P | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA345144225 rs1275262455 |
28 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA345144200 rs1004441593 |
28 | K>N | No |
TOPMed gnomAD ClinGen |
|
|
CA1441437 rs776883496 |
29 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs531385963 CA1441433 |
30 | A>P | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs531385963 CA1441435 |
30 | A>S | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs531385963 CA1441434 |
30 | A>T | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
CA1441432 rs745685235 |
30 | A>V | No |
ExAC TOPMed gnomAD ClinGen |
|
|
rs377552351 CA1441429 |
31 | P>L | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
rs562257505 CA1441430 |
31 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1273784037 CA345144099 |
33 | T>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
TOPMed ClinGen NCI-TCGA |
|
rs200220156 CA345144088 |
33 | T>I | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs200220156 CA1441426 |
33 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA345144076 rs1190544502 |
34 | G>D | No |
ClinGen gnomAD |
|
|
rs750944754 CA345144068 |
35 | G>C | No |
ExAC TOPMed gnomAD ClinGen |
|
|
rs1201973529 CA345144064 |
35 | G>D | No |
ClinGen gnomAD |
|
|
COSM1209731 rs750944754 CA1441423 |
35 | G>S | large_intestine [Cosmic] | No |
ExAC TOPMed gnomAD ClinGen cosmic curated |
|
rs372584075 CA1441420 |
36 | V>A | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA1441421 rs372584075 |
36 | V>E | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA345144050 rs372584075 |
36 | V>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1441422 rs779185305 |
36 | V>M | No |
ExAC gnomAD ClinGen |
|
| TCGA novel | 37 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1441418 rs201151997 |
37 | K>Q | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs752725358 CA1441417 |
37 | K>R | No |
ExAC ClinGen |
|
|
rs774337671 CA1441415 |
38 | K>M | No |
ClinGen ExAC gnomAD |
|
|
rs774337671 CA1441414 |
38 | K>R | No |
ExAC gnomAD ClinGen |
|
|
rs1275672291 CA345143983 |
39 | P>Q | No |
gnomAD ClinGen |
|
|
rs1334201761 CA345143990 |
39 | P>T | No |
gnomAD ClinGen |
|
|
rs1177161942 CA345143964 |
40 | H>R | No |
gnomAD ClinGen |
|
|
CA1441412 rs762754764 |
40 | H>Y | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441410 rs769534943 |
41 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769534943 CA345143928 |
41 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769534943 CA345143931 |
41 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441406 rs766963994 |
42 | Y>* | No |
ExAC gnomAD ClinGen |
|
|
CA1441407 rs144390696 |
42 | Y>C | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA345143865 rs1484326215 |
43 | R>G | No |
TOPMed gnomAD ClinGen |
|
|
CA345143854 rs1238300074 |
43 | R>L | No |
gnomAD ClinGen |
|
|
CA345143849 rs1435843313 |
44 | P>A | No |
gnomAD ClinGen |
|
|
CA345143847 rs1466119123 |
44 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
CA345143823 rs1358711210 |
45 | G>D | No |
gnomAD ClinGen |
|
|
rs746429747 CA1441404 |
45 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs746429747 CA1441405 |
45 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA345143815 rs1358711210 |
45 | G>V | No |
ClinGen gnomAD |
|
|
CA1441402 COSM1209732 rs757564822 |
46 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ExAC gnomAD ClinGen cosmic curated NCI-TCGA |
|
rs757564822 CA345143784 |
46 | T>R | No |
ClinGen ExAC gnomAD |
|
|
CA345143760 rs777992092 |
47 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA1441400 rs777992092 |
47 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs756376731 CA1441399 |
48 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs752880989 CA1441398 |
49 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441397 rs767647652 |
50 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA345143639 rs1161706855 |
51 | E>G | No |
ClinGen gnomAD |
|
|
rs201904037 CA1441396 |
53 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1177664534 CA345143588 |
53 | R>S | No |
gnomAD ClinGen |
|
|
rs751462450 CA1441395 |
54 | R>G | No |
ExAC gnomAD ClinGen |
|
|
CA1441394 rs138699472 |
54 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA38747850 rs867805240 |
55 | Y>* | No |
Ensembl ClinGen |
|
|
CA1441393 rs199974633 |
55 | Y>C | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
CA345143491 rs1205621529 |
56 | Q>* | No |
gnomAD ClinGen |
|
|
CA1441392 rs773014431 |
56 | Q>R | No |
ExAC gnomAD ClinGen |
|
|
CA345143458 rs910118851 |
57 | K>T | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 57 | K>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs910118851 CA38747849 |
57 | K>M | No |
TOPMed gnomAD ClinGen |
|
|
CA345143406 rs1321774460 |
58 | S>C | No |
gnomAD ClinGen |
|
|
rs1321774460 CA345143402 |
58 | S>F | No |
gnomAD ClinGen |
|
|
rs776419197 CA1441389 |
59 | T>A | No |
ExAC gnomAD ClinGen |
|
|
rs768341466 CA1441388 |
59 | T>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ExAC gnomAD ClinGen NCI-TCGA |
|
CA1441386 rs145767549 |
61 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749735656 CA1441384 |
62 | L>P | No |
ExAC gnomAD ClinGen |
|
|
CA345143305 rs2230656 |
63 | I>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1441380 rs201294185 |
64 | R>C | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
CA1441381 rs201294185 |
64 | R>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA345143289 rs1350535396 |
64 | R>L | No |
gnomAD ClinGen |
|
|
CA38747794 rs777329716 |
65 | K>T | No |
ClinGen Ensembl |
|
|
rs755015645 CA345143250 CA1441378 |
66 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345143266 rs781330802 |
66 | L>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781330802 CA1441379 |
66 | L>V | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA1441376 rs148883841 |
67 | P>L | No |
ESP ExAC gnomAD ClinGen |
|
|
CA1441377 rs201625757 |
67 | P>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA1441374 CA345143213 rs144064739 |
68 | F>L | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
rs148655394 CA1441375 |
68 | F>Y | No |
1000Genomes ExAC TOPMed gnomAD ClinGen |
|
|
CA1441373 rs143341207 |
69 | Q>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1215677963 CA345143202 |
69 | Q>R | No |
ClinGen TOPMed |
|
|
CA1441370 rs143639671 |
70 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1441371 rs143639671 |
70 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1441372 rs761689329 |
70 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ExAC gnomAD ClinGen NCI-TCGA |
|
CA1441368 rs150702519 |
72 | M>T | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinGen |
|
|
rs1222575419 CA345143175 |
72 | M>V | No |
ClinGen gnomAD |
|
|
CA1441366 rs148579172 |
73 | R>C | No |
ESP ExAC TOPMed ClinGen |
|
|
rs148579172 CA1441365 |
73 | R>G | No |
ClinGen ESP ExAC TOPMed |
|
|
rs773691912 COSM281714 CA1441364 |
73 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ExAC TOPMed gnomAD ClinGen cosmic curated NCI-TCGA |
|
CA38747726 rs148579172 |
73 | R>S | No |
ClinGen ESP ExAC TOPMed |
|
|
CA1441361 rs781426750 |
75 | I>M | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA345143109 rs1558465152 |
75 | I>V | No |
Ensembl ClinGen |
|
|
rs369460992 CA1441360 |
76 | A>P | No |
ESP ExAC TOPMed gnomAD ClinGen |
|
|
rs369460992 CA345143087 |
76 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs369460992 COSM3418867 CA345143090 |
76 | A>T | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ESP ExAC TOPMed gnomAD ClinGen cosmic curated NCI-TCGA |
|
rs747179869 CA1441359 |
76 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1574109345 CA345143072 |
77 | Q>* | No |
Ensembl ClinGen |
|
|
rs1000042512 CA38747710 |
77 | Q>P | No |
gnomAD ClinGen |
|
|
rs758405073 CA1441357 |
78 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs780227110 CA1441358 |
78 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA345143031 rs1348423079 |
79 | F>I | No |
ClinGen TOPMed |
|
|
rs750444463 CA1441356 |
79 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA1441354 rs765210579 |
80 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1441353 rs753636972 |
81 | T>I | No |
ExAC gnomAD ClinGen |
|
|
rs753636972 CA1441352 |
81 | T>N | No |
ExAC gnomAD ClinGen |
|
|
rs1202634115 CA345142959 |
82 | D>N | No |
gnomAD ClinGen |
|
|
CA38747683 rs143162679 |
83 | L>M | No |
ESP ExAC TOPMed ClinGen |
|
|
CA38747678 rs143162679 |
83 | L>V | No |
ESP ExAC TOPMed ClinGen |
|
|
CA1441347 rs758908744 |
84 | R>C | No |
ClinGen ExAC |
|
|
CA345142885 rs1235890313 |
84 | R>P | No |
gnomAD ClinGen |
|
|
CA345142852 rs1313254665 |
86 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
rs1482262254 CA345142804 |
88 | S>L | No |
ClinGen TOPMed |
|
|
CA345142815 rs1234923318 |
88 | S>P | No |
TOPMed ClinGen |
|
|
rs748538142 CA1441344 |
92 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs1452175748 CA345142694 |
95 | E>G | No |
ClinGen gnomAD |
|
|
CA1441343 rs777033200 |
95 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA1441342 rs768876038 |
96 | A>S | No |
ExAC gnomAD ClinGen |
|
|
rs747322437 COSM1733939 CA1441341 |
96 | A>V | pancreas [Cosmic] | No |
ExAC gnomAD ClinGen cosmic curated |
|
CA345142663 rs201439742 |
98 | E>* | No |
1000Genomes ExAC gnomAD ClinGen |
|
|
rs201439742 CA1441338 |
98 | E>Q | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1179543240 CA345142656 |
98 | E>V | No |
TOPMed ClinGen |
|
|
rs1574109271 CA345142645 |
99 | S>A | No |
Ensembl ClinGen |
|
|
CA1441337 rs776386373 |
100 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 100 | Y>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA345142595 rs137880828 |
101 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD ClinGen |
|
|
CA1441335 rs779013955 |
102 | V>G | No |
ClinGen ExAC gnomAD |
|
|
CA345142580 rs1390238114 |
102 | V>M | No |
ClinGen gnomAD |
|
|
CA345142517 rs1574109251 |
105 | F>V | No |
ClinGen Ensembl |
|
|
CA38747610 rs1052512820 |
106 | E>D | No |
ClinGen TOPMed |
|
|
CA38747612 rs868727723 |
106 | E>G | No |
Ensembl ClinGen |
|
|
CA345142469 rs1558465033 |
107 | D>G | No |
ClinGen Ensembl |
|
|
CA38747606 rs930162029 |
108 | T>A | No |
TOPMed ClinGen |
|
|
CA345142442 rs777690025 |
108 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA1441331 rs777690025 |
108 | T>S | No |
ExAC gnomAD ClinGen |
|
|
rs759075657 CA1441327 |
109 | N>I | No |
ExAC gnomAD ClinGen |
|
|
CA345142415 rs1245345861 |
109 | N>K | No |
ClinGen gnomAD |
|
|
rs759075657 CA1441329 |
109 | N>S | No |
ExAC gnomAD ClinGen |
|
|
CA1441328 rs759075657 |
109 | N>T | No |
ExAC gnomAD ClinGen |
|
|
rs765855738 CA1441326 |
110 | L>V | No |
ExAC TOPMed gnomAD ClinGen |
|
|
rs1041180422 CA38747551 |
111 | C>R | No |
ClinGen Ensembl |
|
|
CA38747550 rs367745564 |
112 | V>A | No |
ClinGen ESP gnomAD |
|
|
rs374791602 CA1441322 |
113 | I>F | No |
ESP ExAC gnomAD ClinGen |
|
|
CA1441321 rs769122987 |
113 | I>M | No |
ExAC TOPMed gnomAD ClinGen |
|
|
CA345142345 rs1215285009 |
113 | I>T | No |
TOPMed ClinGen |
|
|
rs374791602 CA1441323 |
113 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs372972490 CA1441320 |
114 | H>Y | No |
ClinGen ESP ExAC gnomAD |
|
|
CA38747521 rs935358843 |
115 | A>D | No |
TOPMed gnomAD ClinGen |
|
|
CA345142294 rs935358843 |
115 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs772146691 CA1441319 |
116 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772146691 CA1441318 |
116 | K>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA38747518 COSM3689427 rs545672641 |
117 | R>Q | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
1000Genomes ClinGen cosmic curated NCI-TCGA |
|
COSM1997173 CA1441317 rs368511457 |
117 | R>W | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1418217634 CA345142242 |
119 | T>P | No |
ClinGen gnomAD |
|
|
CA1441314 rs749296189 |
120 | I>V | No |
ExAC gnomAD ClinGen |
|
|
CA1441312 rs755941555 |
121 | M>R | No |
ExAC TOPMed gnomAD ClinGen |
|
|
rs1420061280 CA345142220 |
121 | M>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
gnomAD ClinGen NCI-TCGA |
|
rs1187813441 CA345142197 |
122 | P>L | No |
TOPMed ClinGen |
|
|
rs780917924 CA1441310 |
123 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 124 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs751183313 CA1441308 |
126 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs375937549 CA1441307 |
126 | Q>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA345142118 rs1332403307 |
128 | A>V | No |
ClinGen TOPMed |
|
|
rs1431559822 CA345142109 |
129 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA1441305 rs199657441 |
130 | R>C | No |
ExAC TOPMed gnomAD ClinGen |
|
|
rs146830002 CA1441304 |
130 | R>H | No |
ClinGen ESP ExAC gnomAD |
|
|
rs761134743 CA1441303 |
131 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA1441302 rs775764888 |
132 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ExAC gnomAD ClinGen NCI-TCGA |
|
CA345142080 rs775764888 |
132 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA345142076 rs1228332830 |
132 | R>L | No |
TOPMed ClinGen |
|
|
rs759900631 CA1441300 |
133 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA345142057 rs1487311934 |
134 | E>K | No |
TOPMed gnomAD ClinGen |
|
|
CA345142041 rs1277450211 |
135 | R>Q | No |
TOPMed ClinGen |
|
|
rs1163396027 CA345142028 |
136 | A>V | No |
gnomAD ClinGen |
|
|
rs774697896 CA1441299 |
137 | A>W | No |
ClinGen ExAC gnomAD |
No associated diseases with Q16695
1 regional properties for Q16695
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Histone H2A/H2B/H3 | 1 - 132 | IPR007125 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromosome, telomeric region | The end of a linear chromosome, required for the integrity and maintenance of the end. A chromosome telomere usually includes a region of telomerase-encoded repeats the length of which rarely exceeds 20 bp each and that permits the formation of a telomeric loop (T-loop). The telomeric repeat region is usually preceded by a sub-telomeric region that is gene-poor but rich in repetitive elements. Some telomeres only consist of the latter part (for eg. D. melanogaster telomeres). |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleosome | A complex comprised of DNA wound around a multisubunit core and associated proteins, which forms the primary packing unit of DNA into higher order structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| protein heterodimerization activity | Binding to a nonidentical protein to form a heterodimer. |
| structural constituent of chromatin | The action of a molecule that contributes to the structural integrity of chromatin. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| nucleosome assembly | The aggregation, arrangement and bonding together of a nucleosome, the beadlike structural units of eukaryotic chromatin composed of histones and DNA. |
35 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P68428 | Histone H3.2 | Triticum aestivum (Wheat) | PR | |
| Q5E9F8 | H3-3B | Histone H3.3 | Bos taurus (Bovine) | PR |
| P68432 | Histone H3.1 | Bos taurus (Bovine) | PR | |
| P84227 | Histone H3.2 | Bos taurus (Bovine) | PR | |
| P84247 | H3-X | Histone H3.3 | Gallus gallus (Chicken) | PR |
| P84229 | H3-VIII | Histone H3.2 | Gallus gallus (Chicken) | PR |
| P02299 | His3 | Histone H3 | Drosophila melanogaster (Fruit fly) | PR |
| Q71H73 | Histone H3.3 | Vitis vinifera (Grape) | PR | |
| P68431 | H3C12 | Histone H3.1 | Homo sapiens (Human) | PR |
| P84243 | H3-3B | Histone H3.3 | Homo sapiens (Human) | PR |
| Q71DI3 | H3C13 | Histone H3.2 | Homo sapiens (Human) | PR |
| P69246 | H3C4 | Histone H3.2 | Zea mays (Maize) | PR |
| P84228 | H3c15 | Histone H3.2 | Mus musculus (Mouse) | PR |
| P68433 | H3c11 | Histone H3.1 | Mus musculus (Mouse) | PR |
| P84244 | H3-3b | Histone H3.3 | Mus musculus (Mouse) | PR |
| Q71LE2 | H3-3A | Histone H3.3 | Sus scrofa (Pig) | PR |
| P84245 | H3-3b | Histone H3.3 | Rattus norvegicus (Rat) | PR |
| O35799 | Hfe | Hereditary hemochromatosis protein homolog | Rattus norvegicus (Rat) | PR |
| Q6LED0 | Histone H3.1 | Rattus norvegicus (Rat) | PR | |
| Q0JCT1 | H3 | Histone H3.3 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAD9 | H3R-21 | Histone H3.2 | Oryza sativa subsp japonica (Rice) | PR |
| Q27490 | his-70 | Histone H3.3-like type 1 | Caenorhabditis elegans | PR |
| Q10453 | his-71 | Histone H3.3 type 1 | Caenorhabditis elegans | PR |
| Q27532 | his-74 | Histone H3.3-like type 2 | Caenorhabditis elegans | PR |
| P08898 | his-2 | Histone H3 | Caenorhabditis elegans | PR |
| Q9FX60 | At1g13370 | Histone H3-like 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FXI7 | MGH3 | Histone H3-like 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FKQ3 | At5g65350 | Histone H3-like 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P59226 | HTR2 | Histone H3.1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LR02 | At1g75600 | Histone H3-like 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P59169 | HTR4 | Histone H3.3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6P823 | TGas113e22.1 | Histone H3.3 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q28D37 | TGas081o10.1 | Histone H3.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6PI20 | h3f3a | Histone H3.3 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| Q4QRF4 | zgc:113984; | Histone H3.2 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARTKQTARK | STGGKAPRKQ | LATKVARKSA | PATGGVKKPH | RYRPGTVALR | EIRRYQKSTE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LLIRKLPFQR | LMREIAQDFK | TDLRFQSSAV | MALQEACESY | LVGLFEDTNL | CVIHAKRVTI |
| 130 | |||||
| MPKDIQLARR | IRGERA |