Q16644
Gene name |
MAPKAPK3 |
Protein name |
MAP kinase-activated protein kinase 3 |
Names |
MAPK-activated protein kinase 3 , MAPKAP kinase 3 , MAPKAP-K3 , MAPKAPK-3 , MK-3 , EC 2.7.11.1 , Chromosome 3p kinase , 3pK |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:7867 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
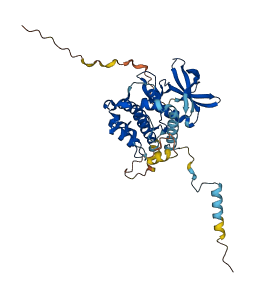
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
4-304 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
186-207 (Activation loop from InterPro)
Target domain |
43-304 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
186-207 (Activation loop from InterPro)
Target domain |
43-304 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Seternes OM et al. (2002) "Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress", Molecular and cellular biology, 22, 6931-45
- Engel K et al. (1998) "Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation", The EMBO journal, 17, 3363-71
Autoinhibited structure

Activated structure

296 variants for Q16644
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000240674 rs886037913 CA10586363 VAR_077085 |
173 | L>P | Patterned macular dystrophy 3 MDPT3; decreased localization to the nucleus [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV002562452 RCV001218305 rs2033320923 |
323 | E>K | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA352961285 rs1204460425 |
2 | D>H | No |
ClinGen gnomAD |
|
|
rs368742045 CA2420130 |
3 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2420131 rs763518219 |
3 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs766895982 CA2420132 |
4 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751937158 CA2420133 |
5 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs372994175 CA74652787 |
9 | Q>* | No |
ClinGen ESP |
|
|
rs955808780 CA352961332 |
9 | Q>P | No |
ClinGen TOPMed |
|
|
CA74652788 rs955808780 |
9 | Q>R | No |
ClinGen TOPMed |
|
|
CA2420135 rs149349769 |
10 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 10 | G>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1183625080 CA352961338 |
10 | G>W | No |
ClinGen gnomAD |
|
|
CA2420139 rs777789251 |
11 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420137 TCGA novel rs147044454 RCV001201570 |
11 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
NCI-TCGA ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA2420138 rs777789251 |
11 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs147044454 CA352961342 |
11 | G>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 12 | P>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1042257886 CA74652855 |
15 | P>L | No |
ClinGen TOPMed |
|
|
rs778722098 CA2420141 |
16 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745672353 CA2420143 |
18 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745672353 CA2420142 |
18 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352961384 rs1238369685 |
19 | P>H | No |
ClinGen gnomAD |
|
|
CA2420144 rs779772290 |
19 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA352961389 rs1286539544 |
20 | G>S | No |
ClinGen gnomAD |
|
|
CA352961394 rs768248070 |
21 | G>* | No |
ClinGen ExAC gnomAD |
|
|
CA352961395 rs776254646 |
21 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420146 rs768248070 |
21 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA2420147 rs776254646 |
21 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352961401 rs1487407777 |
22 | P>L | No |
ClinGen gnomAD |
|
| TCGA novel | 25 | G>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA352961422 rs1436891964 |
26 | G>S | No |
ClinGen gnomAD |
|
|
rs375412266 CA2420150 |
28 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759957245 CA2420152 |
28 | P>L | No |
ClinGen ExAC TOPMed |
|
|
CA2420153 rs759957245 |
28 | P>R | No |
ClinGen ExAC TOPMed |
|
|
VAR_040755 COSM3408768 CA352961435 rs375412266 |
28 | P>S | Variant assessed as Somatic; 0.0 impact. central_nervous_system a glioblastoma multiforme sample; somatic mutation [NCI-TCGA, Cosmic, UniProt] | No |
ClinGen cosmic curated UniProt ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA352961439 rs764208748 |
29 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420156 rs764208748 |
29 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373642329 CA2420154 |
29 | G>R | No |
ClinGen ESP ExAC gnomAD |
|
|
RCV001206489 rs373642329 CA2420155 |
29 | G>W | No |
ClinGen ClinVar ESP ExAC dbSNP gnomAD |
|
|
CA352961443 rs1027886642 |
30 | G>A | No |
ClinGen TOPMed gnomAD |
|
|
CA74652989 rs1027886642 |
30 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA352961451 rs1456669605 |
32 | R>W | No |
ClinGen gnomAD |
|
|
CA2420159 rs368714447 |
35 | K>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 35 | K>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2420160 rs368714447 |
35 | K>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1041696500 CA74653011 |
36 | K>E | No |
ClinGen TOPMed |
|
|
rs1397398299 CA352961489 |
37 | Y>* | No |
ClinGen gnomAD |
|
|
rs1391053347 CA352961492 |
38 | A>P | No |
ClinGen gnomAD |
|
|
rs1391053347 CA352961491 |
38 | A>S | No |
ClinGen gnomAD |
|
|
CA2420162 rs779678577 |
39 | V>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420163 rs746700451 |
40 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420166 rs372238062 |
41 | D>N | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2420167 rs372238062 |
41 | D>Y | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1338275124 CA352961516 |
42 | D>G | No |
ClinGen gnomAD |
|
|
rs78528472 CA74653090 |
42 | D>N | No |
ClinGen gnomAD |
|
|
rs78528472 CA352961513 |
42 | D>Y | No |
ClinGen gnomAD |
|
|
rs1239435558 CA352961563 |
48 | Q>H | No |
ClinGen gnomAD |
|
|
CA352961559 rs1375464774 |
48 | Q>P | No |
ClinGen TOPMed |
|
|
CA352961578 rs1485734849 |
51 | G>D | No |
ClinGen gnomAD |
|
|
rs759919624 CA2420170 |
53 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA2420169 rs759919624 |
53 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1281857290 CA352961606 |
56 | G>S | No |
ClinGen TOPMed |
|
|
CA352961609 rs1479819710 |
56 | G>V | No |
ClinGen gnomAD |
|
| TCGA novel | 60 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs764298483 CA2420173 |
60 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA2420174 rs753977197 |
61 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA352961661 rs1446697428 |
64 | R>Q | No |
ClinGen TOPMed |
|
|
rs1377334631 CA352961660 |
64 | R>W | No |
ClinGen gnomAD |
|
|
rs1302364943 CA352961666 |
65 | R>C | No |
ClinGen gnomAD |
|
|
rs35362731 CA2420176 |
65 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 65 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA352961669 rs750410127 |
66 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420177 rs750410127 |
66 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352961684 RCV001065640 rs1296010021 |
68 | Q>R | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
rs758222981 CA2420178 |
69 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1283225219 CA352925710 |
75 | L>P | No |
ClinGen TOPMed |
|
|
CA352925717 rs1422921375 |
76 | Y>* | No |
ClinGen TOPMed gnomAD |
|
|
CA352925716 rs141282634 |
76 | Y>C | No |
ClinGen ESP gnomAD |
|
|
CA74623782 rs141282634 |
76 | Y>F | No |
ClinGen ESP gnomAD |
|
|
CA352925713 rs1477536792 |
76 | Y>H | No |
ClinGen Ensembl |
|
|
CA2420200 rs752419500 |
81 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs777383707 CA2420202 |
82 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755805527 CA2420201 |
82 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1159952352 CA352925772 |
84 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 86 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs2033150698 RCV001238607 |
86 | D>Y | No |
ClinVar dbSNP |
|
|
CA2420203 rs748745169 |
87 | H>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs748745169 CA352925792 |
87 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA352925790 rs1387729630 |
87 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs575368406 CA2420204 |
91 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
RCV001232934 rs2033151576 |
93 | G>D | No |
ClinVar dbSNP |
|
|
CA352925840 rs1291483185 |
94 | G>A | No |
ClinGen gnomAD |
|
|
RCV001340980 CA2420207 rs143980632 |
94 | G>S | No |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs77366433 CA74623870 |
96 | H>P | No |
ClinGen Ensembl |
|
|
CA352925936 rs1375109467 |
100 | I>T | No |
ClinGen gnomAD |
|
|
rs1188918209 CA352926008 |
104 | Y>F | No |
ClinGen TOPMed |
|
| VAR_040756 | 105 | E>A | an ovarian endometrioid sample; somatic mutation [UniProt] | No | UniProt |
|
rs867974734 CA352926076 CA74623896 |
107 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
CA352926065 rs1559488884 |
107 | M>R | No |
ClinGen Ensembl |
|
|
rs769899324 CA2420210 |
107 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420211 rs773395153 |
109 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs762952384 CA2420212 |
110 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1245151932 CA352926153 |
111 | K>R | No |
ClinGen gnomAD |
|
|
rs200955406 CA2420214 |
112 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA2420215 rs759497061 |
112 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1471567769 CA352926248 |
116 | I>M | No |
ClinGen gnomAD |
|
|
CA352926232 rs1260815780 |
116 | I>V | No |
ClinGen TOPMed |
|
|
CA74623925 rs1032039030 RCV001346405 |
117 | I>T | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1411074539 CA352926284 |
118 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA74623952 rs1029588764 |
120 | C>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1559488925 CA352926350 |
120 | C>Y | No |
ClinGen Ensembl |
|
|
rs1576012847 CA352926619 |
121 | M>I | No |
ClinGen Ensembl |
|
|
CA352926637 rs1258094000 |
122 | E>A | No |
ClinGen gnomAD |
|
|
rs1234361182 CA352926631 |
122 | E>K | No |
ClinGen TOPMed |
|
|
rs1323850753 CA352926660 |
123 | G>S | No |
ClinGen TOPMed |
|
|
rs770936440 CA2420230 |
126 | L>V | No |
ClinGen ExAC gnomAD |
|
|
COSM584380 rs199541398 CA2420233 |
129 | R>S | lung [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs775483656 CA2420234 |
132 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA2420235 rs760401804 |
133 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs763883941 CA2420236 |
133 | R>H | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 134 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs761480203 CA2420238 |
135 | D>N | Variant assessed as Somatic; 0.0001386 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs761480203 CA352927051 |
135 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1282787052 CA352927287 |
142 | E>* | No |
ClinGen gnomAD |
|
|
rs754225374 CA2420266 |
148 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA2420265 rs765824190 COSM236156 |
148 | R>W | lung autonomic_ganglia [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs935316501 CA74625195 |
149 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA352927710 rs935316501 |
149 | D>V | No |
ClinGen TOPMed gnomAD |
|
|
RCV001040833 rs757611175 CA2420267 |
150 | I>T | No |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
|
CA2420268 rs370373095 |
152 | T>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA352927838 rs1410718421 |
153 | A>T | No |
ClinGen gnomAD |
|
|
rs746022177 CA2420269 |
157 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA542863216 rs1458245905 |
158 | H>F | No |
ClinGen gnomAD |
|
|
CA2420271 rs77859836 |
158 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs1386988451 CA352928024 |
158 | H>Y | No |
ClinGen gnomAD |
|
|
rs1324508879 CA352928058 |
159 | S>G | No |
ClinGen gnomAD |
|
|
CA2420272 rs746971211 |
160 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352928197 rs1227782579 |
162 | I>T | No |
ClinGen gnomAD |
|
|
CA2420274 rs776361214 |
162 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA352928217 rs1265975887 |
163 | A>D | No |
ClinGen TOPMed gnomAD |
|
|
CA2420275 rs747984036 |
164 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA74625237 rs757882175 |
165 | R>* | No |
ClinGen gnomAD |
|
|
CA352928273 rs1266182170 |
165 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA352928324 rs1363506925 |
167 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs769508657 CA2420276 |
168 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA352929807 rs1185560872 |
169 | P>A | No |
ClinGen TOPMed |
|
|
CA2420297 rs773801086 |
173 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420299 rs372421280 |
181 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM3696170 rs372421280 CA74626740 |
181 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs944547158 CA74626755 |
181 | A>V | No |
ClinGen Ensembl |
|
|
rs375549664 CA2420301 |
184 | K>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2420302 rs765735423 |
185 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA352930441 rs1255878697 |
186 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
CA2420304 rs763321551 |
193 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA2420306 rs751679355 |
196 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
rs1039317010 CA74626809 |
197 | N>K | No |
ClinGen Ensembl |
|
|
rs2033243949 RCV001236207 |
198 | A>V | No |
ClinVar dbSNP |
|
|
rs755111080 CA74626815 |
202 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420307 rs755111080 |
202 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA74626817 rs766928449 |
207 | Y>D | No |
ClinGen Ensembl |
|
| rs1387018762 | 210 | A>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 210 | A>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1302247089 CA352931108 |
210 | A>T | No |
ClinGen gnomAD |
|
|
rs757096737 CA2420331 |
216 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1328969736 CA352933166 |
222 | S>* | No |
ClinGen gnomAD |
|
|
rs1207253573 CA352933243 |
224 | D>G | No |
ClinGen gnomAD |
|
| TCGA novel | 226 | W>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1488933589 CA352933532 |
229 | G>V | No |
ClinGen gnomAD |
|
|
rs778530686 CA2420332 |
230 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 231 | I>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2420333 rs375875691 |
232 | M>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA2420334 rs375875691 |
232 | M>T | No |
ClinGen ESP ExAC gnomAD |
|
|
rs779765556 CA2420335 |
233 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 233 | Y>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA352933709 rs1411178007 |
235 | L>P | No |
ClinGen gnomAD |
|
|
rs753802960 CA2420348 |
238 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420352 rs758133512 |
243 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs372815253 CA352934052 |
243 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2420351 rs372815253 |
243 | Y>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs779853542 CA2420353 |
244 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA352934099 rs1469284266 |
245 | N>S | No |
ClinGen TOPMed |
|
|
rs1429867795 CA352934114 |
246 | T>A | No |
ClinGen TOPMed |
|
|
rs746597729 RCV001243263 CA2420354 |
246 | T>M | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA74628047 rs780822673 |
247 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754571545 CA2420355 |
247 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA2420356 rs780822673 |
247 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1241745111 CA352934210 |
250 | I>V | No |
ClinGen TOPMed |
|
|
CA74628092 rs975963806 |
252 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs975963806 CA352934272 |
252 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA352934256 rs1212198874 |
252 | P>S | No |
ClinGen gnomAD |
|
|
rs1179935029 CA352934286 |
253 | G>E | No |
ClinGen gnomAD |
|
|
rs1168548018 CA352934368 |
255 | K>M | No |
ClinGen TOPMed |
|
|
rs1168548018 CA352934364 |
255 | K>R | No |
ClinGen TOPMed |
|
|
CA74628118 rs746224997 |
257 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs746224997 CA2420360 |
257 | R>M | No |
ClinGen ExAC gnomAD |
|
|
RCV001223745 rs199901611 CA2420361 |
259 | R>C | No |
ClinGen ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
CA2420362 RCV001349223 rs775712762 |
259 | R>H | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs775712762 CA352934496 |
259 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1360271800 CA352934523 |
261 | G>R | No |
ClinGen gnomAD |
|
|
CA352934564 rs1447672702 |
262 | Q>R | No |
ClinGen gnomAD |
|
|
CA352934603 rs187648751 |
263 | Y>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 263 | Y>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs761866056 CA74628151 |
264 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs372960797 CA2420366 |
264 | G>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA2420367 rs761866056 |
264 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA352934731 rs1576016686 |
267 | N>H | No |
ClinGen Ensembl |
|
|
CA352934766 rs1337584009 |
267 | N>S | No |
ClinGen gnomAD |
|
|
rs1576016700 CA352934901 |
271 | S>A | No |
ClinGen Ensembl |
|
|
rs758137071 CA2420370 |
272 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs750304055 CA2420369 |
272 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA2420371 rs377010404 |
275 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs751301162 CA352935037 |
276 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751301162 CA2420372 |
276 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
VAR_040757 rs56107897 CA74628228 |
276 | D>Y | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs750932578 CA2420396 |
277 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs750932578 CA352935312 |
277 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1351066812 CA352935350 |
279 | Q>K | No |
ClinGen gnomAD |
|
|
CA2420397 rs79652799 RCV001237182 |
282 | R>C | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs747366676 RCV001323969 CA2420399 |
282 | R>H | No |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
|
rs747366676 CA2420400 |
282 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs79652799 CA2420398 |
282 | R>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs748349748 CA2420402 |
283 | L>R | No |
ClinGen ExAC TOPMed |
|
|
rs1484183504 CA352935621 |
288 | D>G | No |
ClinGen TOPMed |
|
|
CA2420403 rs769809042 RCV001321703 |
288 | D>N | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs1254507565 CA352935661 |
289 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1254507565 CA352935658 |
289 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA2420404 rs373989634 |
290 | T>I | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 290 | T>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs774366640 CA2420407 |
297 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA74628702 rs759419783 RCV001351422 CA2420408 |
301 | H>Q | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA2420410 rs752324089 |
302 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs760343500 CA2420411 |
303 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA2420412 rs763562128 |
303 | W>* | Variant assessed as Somatic; 4.62e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| rs957985236 | 305 | N>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1304557436 CA352936523 |
305 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs764674138 CA74628858 |
307 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA2420432 rs764674138 |
307 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1190804603 CA352936779 |
308 | M>T | No |
ClinGen gnomAD |
|
|
rs1391040721 CA352936843 |
309 | V>A | No |
ClinGen gnomAD |
|
|
CA74628868 rs554702337 |
310 | V>M | No |
ClinGen Ensembl |
|
|
CA74628876 rs893709381 |
312 | Q>H | No |
ClinGen Ensembl |
|
|
CA2420436 rs753103323 |
314 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA2420435 rs376026422 |
314 | P>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV001300450 CA74628898 rs868590760 |
315 | L>F | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
CA2420439 rs749474530 RCV001206873 COSM3722473 |
317 | T>M | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated ClinVar ExAC dbSNP gnomAD |
|
CA352937167 rs1364047785 |
318 | A>V | No |
ClinGen gnomAD |
|
|
rs1466112278 CA352937180 |
319 | R>* | No |
ClinGen TOPMed |
|
|
rs779105845 CA352937187 |
319 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
RCV001245148 rs779105845 CA2420441 |
319 | R>Q | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA352937370 rs745848601 |
323 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1477373625 CA352937423 |
324 | D>G | No |
ClinGen TOPMed |
|
|
CA2420443 rs368455680 |
325 | K>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA352937615 rs1262884842 |
327 | H>D | No |
ClinGen gnomAD |
|
|
RCV001352258 rs2033321555 |
328 | W>* | No |
ClinVar dbSNP |
|
|
rs1211001436 CA352937726 |
329 | D>H | No |
ClinGen gnomAD |
|
|
rs768275251 CA352937764 |
330 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA74628934 rs776286044 CA352937805 |
330 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768275251 CA2420446 |
330 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1433672274 CA352937839 |
332 | K>Q | No |
ClinGen gnomAD |
|
|
CA352939571 rs1166410053 |
335 | M>T | No |
ClinGen gnomAD |
|
|
rs1350141172 CA352939599 |
336 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs779191741 CA2420462 |
337 | S>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 337 | S>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA352939658 rs1364236066 RCV001236870 |
338 | A>T | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
RCV001241566 rs750526292 CA2420463 |
338 | A>V | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
CA2420466 rs539746424 |
340 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA2420467 rs768522300 |
341 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA352939942 rs1221769570 |
343 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA2420468 rs557756970 |
343 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1576018619 CA352939946 |
344 | V>I | No |
ClinGen Ensembl |
|
| TCGA novel | 345 | D>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs199560575 CA2420470 |
346 | Y>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA2420469 rs201584585 |
346 | Y>H | No |
ClinGen ExAC gnomAD |
|
|
rs2033344630 RCV001240927 |
346 | Y>Q | No |
ClinVar dbSNP |
|
|
CA352940111 rs1205843411 |
347 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1205843411 CA352940128 |
347 | D>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1232255502 CA352940180 |
348 | Q>K | No |
ClinGen gnomAD |
|
|
rs762542270 CA2420472 |
349 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA352940306 rs1338453711 |
350 | K>N | No |
ClinGen TOPMed |
|
|
CA2420473 rs770298926 |
352 | K>* | No |
ClinGen ExAC gnomAD |
|
|
rs1443688749 CA352940479 |
354 | L>V | No |
ClinGen gnomAD |
|
|
rs761057460 CA2420476 |
355 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1559491747 CA352940541 |
356 | T>I | No |
ClinGen Ensembl |
|
|
rs1432149629 CA352940531 |
356 | T>S | No |
ClinGen gnomAD |
|
|
rs2033345821 RCV001214009 |
357 | S>C | No |
ClinVar dbSNP |
|
|
CA74629390 rs368591672 |
360 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
RCV001063369 CA2420478 rs762190469 |
360 | R>W | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs141897508 CA2420479 |
363 | N>S | No |
ClinGen ESP ExAC gnomAD |
|
|
rs924312729 CA74629401 |
365 | R>K | No |
ClinGen TOPMed |
|
|
CA2420481 rs758524278 |
367 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs779952844 CA2420482 |
369 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1272121660 CA352941091 |
371 | G>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1237906944 CA352941362 |
380 | N>K | No |
ClinGen gnomAD |
|
|
rs1576018745 CA352941352 |
380 | N>T | No |
ClinGen Ensembl |
|
|
rs754885308 CA2420484 |
382 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
1 associated diseases with Q16644
[MIM: 617111]: Macular dystrophy, patterned, 3 (MDPT3)
A form of retinal patterned dystrophy, characterized by retinal pigment epithelium and Bruch's membrane changes resembling a 'dry desert land'. It begins around the age of 30 and progresses to retinitis pigmentosa. MDPT3 inheritance is autosomal dominant. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of retinal patterned dystrophy, characterized by retinal pigment epithelium and Bruch's membrane changes resembling a 'dry desert land'. It begins around the age of 30 and progresses to retinitis pigmentosa. MDPT3 inheritance is autosomal dominant. . Note=The disease is caused by variants affecting the gene represented in this entry.
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent protein serine/threonine kinase activity | Calcium-dependent catalysis of the reactions |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein kinase activity | Calmodulin-dependent catalysis of the reactions |
| MAP kinase kinase activity | Catalysis of the concomitant phosphorylation of threonine (T) and tyrosine (Y) residues in a Thr-Glu-Tyr (TEY) thiolester sequence in a MAP kinase (MAPK) substrate. |
| mitogen-activated protein kinase binding | Binding to a mitogen-activated protein kinase. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| macropinocytosis | An endocytosis process that results in the uptake of liquid material by cells from their external environment by the 'ruffling' of the cell membrane to form heterogeneously sized intracellular vesicles called macropinosomes, which can be up to 5 micrometers in size. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| response to cytokine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| response to lipopolysaccharide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor of a target cell. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
| vascular endothelial growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a vascular endothelial growth factor receptor (VEGFR) on the surface of the target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3SYZ2 | MAPKAPK3 | MAP kinase-activated protein kinase 3 | Bos taurus (Bovine) | SS |
| P49071 | MAPk-Ak2 | MAP kinase-activated protein kinase 2 | Drosophila melanogaster (Fruit fly) | SS |
| Q8IW41 | MAPKAPK5 | MAP kinase-activated protein kinase 5 | Homo sapiens (Human) | SS |
| P49137 | MAPKAPK2 | MAP kinase-activated protein kinase 2 | Homo sapiens (Human) | EV |
| Q16566 | CAMK4 | Calcium/calmodulin-dependent protein kinase type IV | Homo sapiens (Human) | SS |
| O54992 | Mapkapk5 | MAP kinase-activated protein kinase 5 | Mus musculus (Mouse) | EV |
| P49138 | Mapkapk2 | MAP kinase-activated protein kinase 2 | Mus musculus (Mouse) | EV |
| Q3UMW7 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Mus musculus (Mouse) | SS |
| Q66H84 | Mapkapk3 | MAP kinase-activated protein kinase 3 | Rattus norvegicus (Rat) | SS |
| Q965G5 | mak-2 | MAP kinase-activated protein kinase mak-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDGETAEEQG | GPVPPPVAPG | GPGLGGAPGG | RREPKKYAVT | DDYQLSKQVL | GLGVNGKVLE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CFHRRTGQKC | ALKLLYDSPK | ARQEVDHHWQ | ASGGPHIVCI | LDVYENMHHG | KRCLLIIMEC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MEGGELFSRI | QERGDQAFTE | REAAEIMRDI | GTAIQFLHSH | NIAHRDVKPE | NLLYTSKEKD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AVLKLTDFGF | AKETTQNALQ | TPCYTPYYVA | PEVLGPEKYD | KSCDMWSLGV | IMYILLCGFP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PFYSNTGQAI | SPGMKRRIRL | GQYGFPNPEW | SEVSEDAKQL | IRLLLKTDPT | ERLTITQFMN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HPWINQSMVV | PQTPLHTARV | LQEDKDHWDE | VKEEMTSALA | TMRVDYDQVK | IKDLKTSNNR |
| 370 | 380 | ||||
| LLNKRRKKQA | GSSSASQGCN | NQ |