Q15811
Gene name |
ITSN1 (ITSN, SH3D1A) |
Protein name |
Intersectin-1 |
Names |
SH3 domain-containing protein 1A , SH3P17 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6453 |
EC number |
|
Protein Class |
RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR AT 64C, ISOFORM A (PTHR46006) |
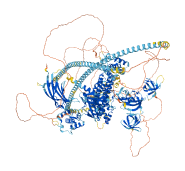
Descriptions
Intersectin 1 is a guanine nucleotide exchange factor (GEF) protein that indirectly coordinates endocytic membrane traffic with the actin assembly machinery. The GEF activity of Intersectin 1 is mediated by its short SH3-DH domain linker, rather than by its SH3 domains. The specific amino acid W1221 within this linker is a key residue for establishing the inhibitory interaction. This mechanism serves to prevent the unregulated activation of the GEF, thereby conserving cellular resources and ensuring precise control of downstream signaling pathways. As SH3 domains are not involved in the autoinhibition, the interaction with the proline-rich region of N-WASP is unable to release the autoinhibition. Although the mechanism by which the autoinhibition is relieved remains unidentified, the activity of Intersectin 1 may be alleviated by transient interactions with binding partners that could mask W1221.
Autoinhibitory domains (AIDs)
Target domain |
1229-1577 (DH-PH domains) |
Relief mechanism |
Partner binding |
Assay |
Structural analysis, Mutagenesis experiment |
Target domain |
741-803 (SH3A domain) |
Relief mechanism |
|
Assay |
|
Target domain |
741-803 (SH3A domain) |
Relief mechanism |
PTM |
Assay |
|
Target domain |
1237-1423 (DH domain) |
Relief mechanism |
Partner binding, Ligand binding |
Assay |
|
Accessory elements
No accessory elements
References
- Kintscher C et al. (2010) "Autoinhibition of GEF activity in Intersectin 1 is mediated by the short SH3-DH domain linker", Protein science : a publication of the Protein Society, 19, 2164-74
- Gerth F et al. (2019) "Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain", Structure (London, England : 1993), 27, 977-987.e5
- Ahmad KF et al. (2010) "The minimal autoinhibited unit of the guanine nucleotide exchange factor intersectin", PloS one, 5, e11291
- Murayama K et al. (2007) "Crystal structure of the rac activator, Asef, reveals its autoinhibitory mechanism", The Journal of biological chemistry, 282, 4238-4242
Autoinhibited structure

Activated structure

12 structures for Q15811
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1KI1 | X-ray | 230 A | B/D | 1229-1581 | PDB |
| 2KGR | NMR | - | A | 210-312 | PDB |
| 2KHN | NMR | - | A | 1-111 | PDB |
| 3FIA | X-ray | 145 A | A | 1-111 | PDB |
| 3QBV | X-ray | 265 A | B/D | 1229-1579 | PDB |
| 4IIM | X-ray | 180 A | A/B | 916-970 | PDB |
| 5HZI | X-ray | 260 A | A/B | 1230-1580 | PDB |
| 5HZJ | X-ray | 260 A | A/B | 1230-1580 | PDB |
| 5HZK | X-ray | 330 A | B/D | 1230-1580 | PDB |
| 6GBU | X-ray | 344 A | B/D/F/H | 1074-1138 | PDB |
| 6H5T | X-ray | 169 A | A/B | 741-840 | PDB |
| AF-Q15811-F1 | Predicted | AlphaFoldDB |
1465 variants for Q15811
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs2067451408 RCV001823688 |
119 | G>D | Nephrotic syndrome [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs763242335 RCV001266623 |
731 | P>S | Inborn genetic diseases [ClinVar] | Yes |
ClinVar ExAC dbSNP gnomAD |
|
rs755276554 RCV000210640 CA358132 |
743 | V>L | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
| rs1219309908 | 4 | F>L | No |
TOPMed gnomAD |
|
| rs959806083 | 5 | P>L | No | TOPMed | |
|
rs1277069165 COSM1211181 |
6 | T>A | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs868634156 | 6 | T>I | No | gnomAD | |
| rs1484793420 | 7 | P>H | No | gnomAD | |
| rs770736261 | 8 | F>Y | No |
ExAC gnomAD |
|
| COSM71227 | 9 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs143930451 | 9 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs376954232 | 10 | G>D | No |
ESP ExAC gnomAD |
|
| rs1393630174 | 13 | D>A | No |
TOPMed gnomAD |
|
| rs2065461362 | 14 | I>T | No | Ensembl | |
| rs748576165 | 17 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs939799478 | 19 | V>I | No | TOPMed | |
| rs2065461852 | 20 | E>K | No | Ensembl | |
|
COSM1030395 rs201302056 |
23 | A>V | Variant assessed as Somatic; MODERATE impact. endometrium bone [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1345761302 | 26 | D>E | No | gnomAD | |
| rs541450123 | 26 | D>H | No |
1000Genomes ExAC gnomAD |
|
| rs774349088 | 28 | Q>K | No |
ExAC gnomAD |
|
| rs1369976412 | 30 | H>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs772037712 | 31 | S>I | No |
ExAC gnomAD |
|
| TCGA novel | 32 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065463452 | 34 | P>S | No | TOPMed | |
| rs919617743 | 35 | I>M | No |
TOPMed gnomAD |
|
| rs760484132 | 35 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2065463683 | 40 | T>A | No | Ensembl | |
| rs1273294608 | 42 | D>E | No | gnomAD | |
| rs563383929 | 43 | Q>R | No | gnomAD | |
| rs2065566437 | 44 | A>T | No | Ensembl | |
| rs776357486 | 46 | N>D | No |
ExAC gnomAD |
|
| COSM1030396 | 48 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1283492787 | 50 | Q>* | No | Ensembl | |
| rs764851463 | 50 | Q>L | No |
ExAC gnomAD |
|
| rs746959118 | 50 | Q>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746959118 | 50 | Q>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1189528805 | 51 | S>F | No | gnomAD | |
| rs1601839500 | 51 | S>P | No | Ensembl | |
| rs1371201178 | 52 | G>E | No | gnomAD | |
| rs1371201178 | 52 | G>V | No | gnomAD | |
|
COSM3550423 COSM3550422 COSM3550424 |
53 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1601839586 | 53 | L>S | No | Ensembl | |
| rs1168244844 | 54 | P>L | No | gnomAD | |
| rs2065568623 | 54 | P>S | No | Ensembl | |
| rs1371448009 | 56 | P>S | No | gnomAD | |
| TCGA novel | 57 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1311201242 | 58 | L>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756633876 | 62 | W>* | No |
ExAC TOPMed gnomAD |
|
| rs753267635 | 62 | W>R | No |
ExAC gnomAD |
|
| rs866433980 | 63 | A>T | No | Ensembl | |
| rs762114380 | 65 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1343660916 | 66 | D>N | No | TOPMed | |
| rs765375043 | 67 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs765375043 | 67 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1569054625 | 68 | N>S | No |
TOPMed gnomAD |
|
| rs1337321604 | 72 | R>G | No | TOPMed | |
| rs750655480 | 72 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs754272885 | 76 | V>M | No |
TOPMed gnomAD |
|
| rs758689244 | 80 | I>L | No |
ExAC TOPMed gnomAD |
|
|
rs758689244 COSM4134905 COSM4134906 COSM4134904 |
80 | I>V | thyroid [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| TCGA novel | 81 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754899386 | 85 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1433950863 | 86 | K>R | No |
TOPMed gnomAD |
|
| rs1433950863 | 86 | K>T | No |
TOPMed gnomAD |
|
| rs2066418190 | 90 | Q>K | No | Ensembl | |
| rs983870457 | 92 | Y>C | No |
TOPMed gnomAD |
|
| rs1403248326 | 93 | Q>H | No | TOPMed | |
| rs1372506949 | 99 | P>H | No |
TOPMed gnomAD |
|
|
TCGA novel rs769493415 |
100 | P>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA ExAC TOPMed gnomAD |
| rs1569054925 | 100 | P>S | No |
TOPMed gnomAD |
|
| rs748942101 | 104 | Q>R | No |
ExAC gnomAD |
|
| rs2147258849 | 105 | Q>* | No | Ensembl | |
| rs770743433 | 106 | P>T | No |
ExAC gnomAD |
|
| rs1388138645 | 107 | V>F | No |
TOPMed gnomAD |
|
| rs1388138645 | 107 | V>I | No |
TOPMed gnomAD |
|
| rs2066420653 | 109 | I>F | No | TOPMed | |
| rs769166042 | 109 | I>M | No |
ExAC gnomAD |
|
| rs1051601977 | 110 | S>P | No | Ensembl | |
| rs777007166 | 111 | S>C | No |
ExAC gnomAD |
|
|
rs201039916 COSM1413910 |
112 | A>T | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM224071 rs1332542126 |
113 | P>S | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1332542126 | 113 | P>T | No |
TOPMed gnomAD |
|
| rs570083738 | 116 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1436937696 | 116 | G>V | No | gnomAD | |
| rs936959859 | 117 | M>L | No |
TOPMed gnomAD |
|
| rs778568026 | 117 | M>T | No |
ExAC gnomAD |
|
| rs936959859 | 117 | M>V | No |
TOPMed gnomAD |
|
| rs2067451138 | 118 | G>E | No | TOPMed | |
| rs2067451408 | 119 | G>V | No | Ensembl | |
| rs150191551 | 121 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150191551 | 121 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200423943 | 125 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200423943 | 125 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1314522112 | 125 | P>T | No | gnomAD | |
| rs773716558 | 126 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs749601675 | 127 | T>I | No |
ExAC gnomAD |
|
| rs749601675 | 127 | T>R | No |
ExAC gnomAD |
|
| rs1465823122 | 128 | A>S | No |
TOPMed gnomAD |
|
| rs1465823122 | 128 | A>T | No |
TOPMed gnomAD |
|
| rs1161914635 | 129 | V>I | No |
TOPMed gnomAD |
|
| rs771275208 | 130 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs771275208 | 130 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs767827917 | 132 | V>L | No |
ExAC gnomAD |
|
| rs767827917 | 132 | V>M | No |
ExAC gnomAD |
|
| rs2067455620 | 133 | P>L | No | TOPMed | |
| rs775378886 | 133 | P>S | No |
ExAC gnomAD |
|
| rs113818794 | 134 | M>K | No | Ensembl | |
| rs113818794 | 134 | M>T | No | Ensembl | |
| rs372092044 | 134 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2147467504 | 135 | G>E | No | Ensembl | |
| rs1425676660 | 137 | I>V | No |
TOPMed gnomAD |
|
| rs2067456852 | 138 | P>L | No | TOPMed | |
| COSM1030398 | 138 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs199546978 | 139 | V>F | No |
1000Genomes ExAC gnomAD |
|
| rs1458649658 | 141 | G>R | No | gnomAD | |
| rs901616099 | 142 | M>I | No | Ensembl | |
| rs2067457754 | 142 | M>T | No | Ensembl | |
| rs1465413447 | 143 | S>C | No |
TOPMed gnomAD |
|
| rs2067458492 | 145 | T>I | No | Ensembl | |
| rs375382856 | 147 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3841825 COSM3841823 COSM3841824 |
148 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1269793562 | 148 | S>F | No |
TOPMed gnomAD |
|
| rs528600328 | 150 | V>I | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 151 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1315404273 COSM365784 |
151 | P>S | lung [Cosmic] | No |
cosmic curated Ensembl |
| rs2067459859 | 152 | T>I | No | Ensembl | |
| rs1328376656 | 153 | A>T | No | TOPMed | |
| rs1428416450 | 154 | A>S | No |
TOPMed gnomAD |
|
| rs899089428 | 155 | V>M | No | TOPMed | |
| rs757940900 | 156 | P>H | No |
ExAC TOPMed gnomAD |
|
| rs757940900 | 156 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs757940900 | 156 | P>R | No |
ExAC TOPMed gnomAD |
|
|
rs1340587657 COSM3550425 COSM3550426 COSM3550427 |
156 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs754500876 | 157 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs749794095 | 157 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs754500876 | 157 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs754500876 | 157 | P>T | No |
ExAC TOPMed gnomAD |
|
|
rs751926247 COSM4336855 COSM1413911 COSM4336854 |
158 | L>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs2067461709 | 158 | L>V | No |
TOPMed gnomAD |
|
|
COSM2844089 COSM4336857 COSM4336856 |
158 | L>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3748765 rs2147468763 COSM3748763 |
159 | A>T | stomach [Cosmic] | No |
cosmic curated Ensembl |
| rs775859329 | 161 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs768841848 | 162 | A>S | No |
ExAC gnomAD |
|
| rs776915893 | 163 | P>L | No |
ExAC gnomAD |
|
| rs1569093972 | 163 | P>S | No | Ensembl | |
| rs2067462810 | 164 | P>A | No | Ensembl | |
| rs1412673800 | 164 | P>R | No | gnomAD | |
| rs2067463387 | 165 | V>A | No | gnomAD | |
| rs1443009054 | 166 | I>K | No | TOPMed | |
| rs1473521662 | 166 | I>M | No |
TOPMed gnomAD |
|
| rs1443009054 | 166 | I>T | No | TOPMed | |
| rs2067463775 | 167 | Q>* | No | TOPMed | |
| rs765244876 | 167 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs750145035 | 168 | P>S | No |
ExAC TOPMed gnomAD |
|
| COSM1495183 | 169 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765893804 | 171 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs765893804 | 171 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1361246792 | 171 | A>V | No |
TOPMed gnomAD |
|
| rs2067464746 | 173 | A>V | No | gnomAD | |
| rs754695322 | 174 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1448746913 | 177 | A>T | No | TOPMed | |
| rs1569097820 | 178 | T>A | No | Ensembl | |
| rs2067575768 | 180 | P>Q | No | TOPMed | |
| rs981518055 | 181 | K>E | No |
TOPMed gnomAD |
|
| rs981518055 | 181 | K>Q | No |
TOPMed gnomAD |
|
| rs369810887 | 183 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs2067576579 | 184 | S>F | No | gnomAD | |
| rs2067576805 | 186 | S>R | No | TOPMed | |
| rs1227209877 | 187 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2067577281 | 188 | S>A | No | Ensembl | |
|
COSM1030400 rs2067577489 |
188 | S>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1274827765 | 189 | G>A | No | gnomAD | |
| rs2067577915 | 191 | G>R | No |
TOPMed gnomAD |
|
| rs750927643 | 192 | S>L | No |
ExAC gnomAD |
|
| rs780534584 | 193 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1348764255 | 195 | N>S | No | TOPMed | |
| rs2067579238 | 196 | T>A | No | Ensembl | |
| rs544678761 | 197 | K>R | No |
1000Genomes ExAC gnomAD |
|
|
COSM6094709 COSM6094708 COSM6094707 |
197 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1485863696 | 198 | L>* | No | gnomAD | |
| rs1192284766 | 199 | Q>R | No | gnomAD | |
| rs2147493678 | 201 | A>E | No | Ensembl | |
| TCGA novel | 201 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1317465702 | 201 | A>T | No | TOPMed | |
| rs769907006 | 204 | F>L | No | ExAC | |
| rs773234531 | 206 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs749222997 | 207 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs2067581481 | 207 | A>S | No | Ensembl | |
| rs749222997 | 207 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2067581834 | 208 | S>C | No |
TOPMed gnomAD |
|
| rs2067581834 | 208 | S>G | No |
TOPMed gnomAD |
|
| rs770811360 | 209 | V>I | No |
ExAC gnomAD |
|
| rs774273932 | 210 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1218525115 | 210 | P>S | No |
TOPMed gnomAD |
|
| rs1266197843 | 211 | P>S | No | gnomAD | |
| rs1040815582 | 213 | A>V | No | Ensembl | |
| rs546895300 | 214 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1269773434 | 216 | A>S | No | gnomAD | |
| rs1013208908 | 217 | V>I | No | gnomAD | |
| rs1013208908 | 217 | V>L | No | gnomAD | |
| rs897818273 | 218 | P>S | No | gnomAD | |
| rs1343517714 | 222 | R>K | No | gnomAD | |
| rs1266877010 | 223 | L>R | No |
TOPMed gnomAD |
|
| TCGA novel | 225 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2067834828 | 226 | R>S | No | Ensembl | |
| rs1294147621 | 230 | N>D | No |
TOPMed gnomAD |
|
| rs1288719261 | 232 | H>Q | No | gnomAD | |
| rs1201034876 | 232 | H>R | No | gnomAD | |
| rs1410446903 | 233 | D>E | No | gnomAD | |
| rs2067835731 | 235 | T>S | No | Ensembl | |
| rs1414453227 | 236 | M>I | No | TOPMed | |
| rs775212836 | 237 | S>T | No |
ExAC gnomAD |
|
| rs1391281976 | 238 | G>R | No | gnomAD | |
| rs747001526 | 239 | H>Y | No |
TOPMed gnomAD |
|
| rs1344557389 | 241 | T>A | No | TOPMed | |
| rs1489993661 | 243 | P>R | No |
TOPMed gnomAD |
|
| rs768289000 | 243 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs761617958 | 246 | R>G | No |
ExAC gnomAD |
|
| rs1227567678 | 246 | R>T | No | gnomAD | |
| rs562662000 | 247 | T>A | No | TOPMed | |
| rs769426337 | 247 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs772892155 | 248 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs200049832 | 251 | Q>H | No |
1000Genomes ExAC gnomAD |
|
| rs1199209058 | 251 | Q>R | No | gnomAD | |
| rs1341559336 | 255 | P>L | No | gnomAD | |
| rs767936019 | 256 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs1325623468 | 257 | A>G | No | TOPMed | |
| TCGA novel | 257 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2068364021 | 258 | Q>R | No | gnomAD | |
| rs558812785 | 262 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs772559655 | 264 | N>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3550428 COSM3550429 COSM3550430 |
265 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2068689186 | 266 | S>Y | No | TOPMed | |
| rs2068689412 | 268 | I>V | No |
TOPMed gnomAD |
|
| TCGA novel | 270 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775871104 | 270 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2068690156 | 271 | D>E | No | gnomAD | |
| rs761300104 | 271 | D>G | No |
ExAC TOPMed |
|
| TCGA novel | 273 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 274 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754239354 | 275 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1364810311 | 276 | A>P | No | gnomAD | |
| rs1436084537 | 280 | I>T | No |
TOPMed gnomAD |
|
| rs2068691235 | 282 | A>T | No | Ensembl | |
| COSM1030402 | 283 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765466442 | 283 | M>R | No |
ExAC gnomAD |
|
| rs2068691733 | 283 | M>V | No | Ensembl | |
| rs1602100643 | 284 | H>P | No | Ensembl | |
| rs1602100643 | 284 | H>R | No | Ensembl | |
| rs2068692502 | 285 | L>V | No | TOPMed | |
| rs1006326896 | 287 | D>E | No |
TOPMed gnomAD |
|
| rs1326230839 | 288 | V>I | No | gnomAD | |
| rs920377572 | 290 | M>I | No | gnomAD | |
| rs1602100784 | 292 | G>D | No | Ensembl | |
| rs2068693823 | 292 | G>R | No | Ensembl | |
| rs1569131960 | 296 | P>L | No | Ensembl | |
| rs766538916 | 296 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1244562242 | 301 | P>L | No |
TOPMed gnomAD |
|
| rs747981470 | 302 | E>A | No |
ExAC TOPMed gnomAD |
|
| rs747981470 | 302 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs367655089 | 302 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs747981470 | 302 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1471561364 | 307 | S>C | No | gnomAD | |
| rs2068696469 | 307 | S>P | No | TOPMed | |
| rs755791159 | 309 | R>K | No |
ExAC gnomAD |
|
| rs756971499 | 311 | V>A | No |
ExAC gnomAD |
|
|
COSM3550431 COSM175181 COSM3550432 rs1355286586 |
312 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1355286586 | 312 | R>G | No | gnomAD | |
|
COSM1211179 rs778262651 |
312 | R>Q | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs376858445 | 313 | S>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs376858445 | 313 | S>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1203759115 | 315 | S>I | No | gnomAD | |
| rs769110337 | 316 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs2068828702 | 317 | I>T | No |
TOPMed gnomAD |
|
| rs368797638 | 317 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs773608079 | 323 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs773608079 | 323 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1387263667 | 324 | S>C | No | gnomAD | |
| rs1387263667 | 324 | S>F | No | gnomAD | |
| rs763065231 | 325 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1312208229 | 326 | D>G | No | TOPMed | |
| rs766649165 | 326 | D>Y | No |
ExAC gnomAD |
|
| rs1412126844 | 327 | Q>E | No | gnomAD | |
| rs1332693153 | 328 | R>K | No |
TOPMed gnomAD |
|
| rs1453770414 | 330 | P>T | No |
TOPMed gnomAD |
|
| rs2068831463 | 331 | E>G | No |
TOPMed gnomAD |
|
| rs1362538275 | 332 | E>K | No | gnomAD | |
| rs2068832327 | 333 | P>Q | No | Ensembl | |
| rs759669553 | 333 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs759669553 | 333 | P>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 334 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752664591 | 334 | V>I | No |
ExAC gnomAD |
|
| rs967588148 | 338 | E>V | No | TOPMed | |
| rs1184959164 | 339 | Q>R | No | gnomAD | |
|
COSM3423929 COSM3423930 COSM3423928 |
344 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760758707 | 346 | L>S | No |
ExAC gnomAD |
|
| TCGA novel | 347 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 347 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2068833447 | 347 | P>R | No | TOPMed | |
| rs2069206864 | 349 | T>A | No | TOPMed | |
| rs1408030128 | 352 | D>G | No |
TOPMed gnomAD |
|
| rs2147786663 | 354 | K>E | No | Ensembl | |
|
COSM4295812 COSM4295811 rs920288267 COSM1030403 |
360 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs770995793 | 360 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs770995793 | 360 | R>L | No |
ExAC gnomAD |
|
| rs1265992809 | 361 | G>C | No | gnomAD | |
| rs779765019 | 366 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs749185030 | 367 | K>R | No |
ExAC TOPMed gnomAD |
|
|
rs775749120 COSM1030404 |
368 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC gnomAD |
| rs980737897 | 371 | A>T | No | Ensembl | |
| TCGA novel | 372 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 374 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1196805274 | 375 | Q>L | No | gnomAD | |
| rs147000769 | 376 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs764147892 | 377 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs768597698 | 377 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs768597698 COSM334348 |
377 | R>L | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| COSM725237 | 381 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2069212919 | 381 | E>Q | No | Ensembl | |
|
rs764739038 COSM329433 |
382 | R>C | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
rs750087030 COSM261471 |
382 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs750087030 | 382 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs1449015629 | 383 | L>M | No | gnomAD | |
| rs143580796 | 386 | L>V | No |
ESP TOPMed gnomAD |
|
| rs375451100 | 387 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs200196886 | 388 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1384612670 | 388 | R>W | No |
1000Genomes gnomAD |
|
| rs544261869 | 389 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1569145903 | 390 | E>A | No | Ensembl | |
| rs2069216311 | 390 | E>D | No | TOPMed | |
| rs771317373 | 392 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2069217100 | 395 | E>K | No | TOPMed | |
| rs1261569452 | 396 | R>C | No |
TOPMed gnomAD |
|
| rs779207769 | 396 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779207769 | 396 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs772469526 | 398 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs533840234 | 398 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs533840234 | 398 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1250189865 | 399 | Q>E | No | gnomAD | |
| rs2069218867 | 400 | E>K | No | Ensembl | |
| rs2069219379 | 401 | Q>E | No | gnomAD | |
| rs2069219680 | 401 | Q>R | No | TOPMed | |
| rs768410170 | 402 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2069220383 | 403 | R>C | No | TOPMed | |
| rs776643469 | 403 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs776643469 | 403 | R>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 404 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs945492858 | 404 | K>R | No | Ensembl | |
| rs201073854 | 406 | Q>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1569146163 RCV000678374 |
408 | E>* | No |
ClinVar Ensembl dbSNP |
|
| rs1442707240 | 412 | Q>R | No | gnomAD | |
| rs2069223081 | 414 | E>K | No | TOPMed | |
| rs762545126 | 415 | K>Q | No | ExAC | |
| rs766054300 | 416 | Q>E | No |
ExAC TOPMed gnomAD |
|
| rs777736654 | 417 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM282031 rs751006736 |
417 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs766997437 | 419 | L>I | No |
ExAC gnomAD |
|
|
rs2147788557 RCV001579322 |
420 | E>* | No |
ClinVar Ensembl dbSNP |
|
| rs2069225498 | 421 | R>Q | No | gnomAD | |
| rs1308860031 | 421 | R>W | No |
TOPMed gnomAD |
|
| rs373488298 | 425 | E>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs754360446 | 425 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs754360446 | 425 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs2069226515 | 427 | R>K | No | TOPMed | |
| rs1226317955 | 428 | R>G | No |
TOPMed gnomAD |
|
| rs903875641 | 428 | R>K | No | Ensembl | |
| rs1291243061 | 429 | K>R | No | gnomAD | |
| rs2069227826 | 431 | I>V | No | TOPMed | |
| rs533098631 | 433 | R>K | No | 1000Genomes | |
| rs2069229342 | 434 | R>* | No | TOPMed | |
| rs551176344 | 435 | E>* | No | 1000Genomes | |
| TCGA novel | 435 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs551176344 | 435 | E>Q | No | 1000Genomes | |
| rs1482499220 | 436 | A>T | No | gnomAD | |
| rs963769471 | 439 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs547639837 | 439 | R>W | No |
1000Genomes ExAC gnomAD |
|
| rs1602154377 | 440 | E>D | No | Ensembl | |
| rs1446067594 | 441 | L>F | No | gnomAD | |
| rs773975976 | 442 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs371447448 | 443 | R>K | No |
ESP TOPMed gnomAD |
|
| rs759248303 | 445 | R>Q | No |
ExAC gnomAD |
|
| rs767027223 | 446 | Q>P | No | ExAC | |
| rs2069465108 | 447 | L>F | No |
TOPMed gnomAD |
|
| rs1357322509 | 449 | W>S | No | gnomAD | |
| rs1337125488 | 451 | R>Q | No | gnomAD | |
| rs757736852 | 451 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2069466169 | 453 | R>Q | No | TOPMed | |
| rs144048618 | 456 | E>D | No |
ESP TOPMed gnomAD |
|
| rs951098593 | 460 | Q>* | No | TOPMed | |
| rs951098593 | 460 | Q>E | No | TOPMed | |
|
COSM3423931 COSM3423933 COSM3423932 |
461 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2069467159 | 461 | R>K | No | TOPMed | |
| TCGA novel | 462 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2069467334 | 462 | N>S | No | TOPMed | |
|
RCV001579323 rs2147816344 |
463 | K>missing | No |
ClinVar dbSNP |
|
| rs2069467523 | 463 | K>R | No | TOPMed | |
| rs780624357 | 466 | E>A | No |
ExAC gnomAD |
|
| COSM459625 | 466 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1203493739 | 467 | D>G | No | gnomAD | |
| rs1264135160 | 468 | I>T | No | gnomAD | |
| rs566065722 | 470 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs566065722 | 470 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs748205911 | 473 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2069470150 | 474 | K>E | No | TOPMed | |
| rs1431279627 | 476 | K>N | No | TOPMed | |
| rs536525393 | 477 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs180901992 | 478 | L>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM5346630 COSM1307754 COSM5346629 |
483 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2069482209 | 486 | N>S | No | TOPMed | |
| rs745684064 | 487 | D>N | No |
ExAC gnomAD |
|
| rs2069482802 | 487 | D>V | No | gnomAD | |
| rs1279326746 | 489 | K>Q | No | gnomAD | |
|
COSM5205900 COSM5205901 COSM5205899 |
489 | K>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1225118460 | 490 | H>Y | No | gnomAD | |
| rs2069483958 | 491 | Q>E | No | Ensembl | |
| rs775020541 | 491 | Q>H | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 493 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1569153072 | 493 | E>G | No | Ensembl | |
| rs2069484733 | 496 | L>F | No |
TOPMed gnomAD |
|
|
COSM4101394 COSM4101396 COSM4101395 |
496 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1358137811 | 497 | Q>K | No | gnomAD | |
| rs1222542120 | 498 | D>A | No | gnomAD | |
| rs2069485496 | 498 | D>N | No | Ensembl | |
| rs1224067066 | 499 | I>L | No |
TOPMed gnomAD |
|
| rs1049465197 | 499 | I>T | No |
TOPMed gnomAD |
|
| rs1482024961 | 500 | R>T | No | gnomAD | |
| rs1411170207 | 502 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs527827314 | 502 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751883177 | 504 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1424144806 | 504 | T>I | No |
TOPMed gnomAD |
|
| rs767980274 | 510 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs759829776 | 510 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs893380850 | 512 | S>N | No |
TOPMed gnomAD |
|
| rs574816046 | 513 | T>I | No |
1000Genomes ExAC gnomAD |
|
| rs1444755848 | 514 | N>D | No | gnomAD | |
| rs777811798 | 514 | N>I | No |
ExAC gnomAD |
|
| rs1381554892 | 515 | K>N | No |
TOPMed gnomAD |
|
| rs2069490452 | 515 | K>Q | No | gnomAD | |
| rs2069490892 | 516 | S>* | No | Ensembl | |
| rs1247155458 | 516 | S>Y | No | gnomAD | |
| rs2069492166 | 517 | R>G | No | Ensembl | |
| rs753939781 | 518 | E>K | No |
ExAC gnomAD |
|
| rs757148859 | 518 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs2069494416 | 519 | L>F | No | Ensembl | |
| rs1195167683 | 519 | L>S | No | gnomAD | |
| rs778828523 | 520 | R>K | No |
ExAC gnomAD |
|
| COSM1030405 | 521 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745620946 | 522 | A>G | No |
ExAC gnomAD |
|
|
COSM4693649 rs779966613 COSM169917 COSM4693648 |
523 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1444225684 | 523 | E>V | No | gnomAD | |
| rs2069495739 | 524 | I>V | No | Ensembl | |
| rs746611473 | 525 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1017738636 | 525 | T>I | No | gnomAD | |
| rs1474434707 | 526 | H>Y | No |
TOPMed gnomAD |
|
| rs2069497005 | 527 | L>P | No | Ensembl | |
| rs901993996 | 527 | L>V | No | TOPMed | |
| rs1397789205 | 529 | Q>R | No |
TOPMed gnomAD |
|
| rs144443158 | 530 | Q>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2147820108 | 530 | Q>P | No | Ensembl | |
| TCGA novel | 534 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1450784424 | 536 | Q>R | No | TOPMed | |
| TCGA novel | 537 | M>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781002829 | 538 | L>F | No |
ExAC gnomAD |
|
| rs998543786 | 539 | G>A | No |
TOPMed gnomAD |
|
| rs747858345 | 540 | R>G | No | ExAC | |
| rs769420583 | 540 | R>S | No |
ExAC gnomAD |
|
| rs373643952 | 541 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2070174418 | 542 | I>T | No | Ensembl | |
| rs1282495880 | 543 | P>L | No | gnomAD | |
| rs746335223 | 544 | E>G | No |
ExAC gnomAD |
|
| rs1207143006 | 546 | Q>R | No | gnomAD | |
| rs374935070 | 547 | I>M | No | gnomAD | |
| rs775681162 | 549 | N>K | No |
ExAC gnomAD |
|
| rs142361441 | 549 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs761035834 | 550 | D>E | No |
ExAC gnomAD |
|
|
COSM419249 rs2070176440 |
551 | Q>R | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs2070176884 | 552 | L>F | No | TOPMed | |
| rs367784015 | 555 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs1197912762 | 556 | Q>E | No | gnomAD | |
| rs2147888370 | 558 | N>S | No | 1000Genomes | |
| rs750566466 | 561 | H>P | No |
ExAC gnomAD |
|
| rs1179001631 | 561 | H>Y | No | gnomAD | |
| rs1433414458 | 563 | D>G | No | TOPMed | |
|
COSM4852837 COSM4852836 COSM4852838 |
563 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139132438 | 565 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs139132438 | 565 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs201440281 | 565 | L>R | No |
1000Genomes ExAC gnomAD |
|
| rs766425642 | 567 | T>I | No | ExAC | |
| rs759585042 | 568 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1360176214 | 569 | K>Q | No | gnomAD | |
| rs1480529222 | 571 | A>D | No |
TOPMed gnomAD |
|
| TCGA novel | 571 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1250079646 | 571 | A>P | No |
TOPMed gnomAD |
|
| rs1250079646 | 571 | A>T | No |
TOPMed gnomAD |
|
| rs1480529222 | 571 | A>V | No |
TOPMed gnomAD |
|
| rs1196716416 | 572 | L>V | No | gnomAD | |
| rs1239673875 | 573 | E>D | No |
TOPMed gnomAD |
|
| rs781625948 | 573 | E>Q | No | Ensembl | |
| rs914731886 | 574 | A>T | No | TOPMed | |
| rs755686300 | 575 | K>R | No |
ExAC gnomAD |
|
|
rs2070234124 TCGA novel RCV001579320 COSM173562 |
576 | E>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated ClinVar NCI-TCGA Ensembl dbSNP |
| rs777491450 | 577 | L>I | No |
ExAC gnomAD |
|
| rs749894224 | 579 | R>L | No |
TOPMed gnomAD |
|
| rs749894224 | 579 | R>Q | No |
TOPMed gnomAD |
|
| rs371785092 | 579 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1372394557 | 580 | Q>E | No | gnomAD | |
| rs1462347653 | 581 | H>P | No | gnomAD | |
| rs2070235481 | 581 | H>Q | No | Ensembl | |
| rs756781441 | 583 | R>Q | No |
ExAC gnomAD |
|
| rs1450733750 | 584 | D>E | No | gnomAD | |
| rs778462764 | 584 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1891697059 | 586 | L>P | No | TOPMed | |
| rs2070236815 | 587 | D>V | No | Ensembl | |
|
COSM3841830 COSM3841829 COSM3841831 |
588 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1352223484 | 589 | V>M | No | TOPMed | |
| rs747485694 | 591 | K>R | No |
ExAC gnomAD |
|
| rs1307405930 | 594 | R>G | No | gnomAD | |
| rs1228689726 | 594 | R>I | No |
TOPMed gnomAD |
|
| rs1228689726 | 594 | R>K | No |
TOPMed gnomAD |
|
| rs1228689726 | 594 | R>T | No |
TOPMed gnomAD |
|
| rs781488270 | 595 | S>T | No |
ExAC TOPMed gnomAD |
|
| COSM1030406 | 596 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1460213851 | 598 | Q>* | No | gnomAD | |
| rs2070239210 | 598 | Q>H | No | Ensembl | |
| rs1182811689 | 599 | E>K | No | gnomAD | |
| rs887041382 | 600 | I>T | No |
TOPMed gnomAD |
|
| rs1050984846 | 600 | I>V | No |
TOPMed gnomAD |
|
| rs769850078 | 601 | D>G | No |
ExAC gnomAD |
|
| rs1003851448 | 604 | N>S | No |
TOPMed gnomAD |
|
| rs1158034478 | 604 | N>Y | No | gnomAD | |
| rs1479636163 | 605 | N>D | No | Ensembl | |
| rs1184233063 | 605 | N>S | No | gnomAD | |
| rs1418783486 | 606 | Q>H | No | gnomAD | |
| rs1429042830 | 607 | L>P | No | gnomAD | |
| rs2070242384 | 608 | K>R | No | Ensembl | |
| rs1218534935 | 609 | E>* | No | TOPMed | |
| rs772020474 | 610 | L>I | No | 1000Genomes | |
|
COSM3550433 COSM3550435 COSM3550434 |
611 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs745953273 | 613 | I>M | No |
ExAC gnomAD |
|
| rs774528531 | 613 | I>T | No |
ExAC gnomAD |
|
| rs1284318009 | 614 | H>Y | No | gnomAD | |
| rs775557166 | 619 | L>R | No |
ExAC gnomAD |
|
| rs1240291912 | 621 | K>N | No |
TOPMed gnomAD |
|
| rs1283061258 | 624 | S>P | No |
TOPMed gnomAD |
|
| rs1031160893 | 625 | M>L | No |
TOPMed gnomAD |
|
|
COSM1534754 COSM6161379 COSM6161381 rs1266460178 COSM6161380 |
625 | M>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1031160893 | 625 | M>V | No |
TOPMed gnomAD |
|
| rs760599859 | 626 | E>D | No |
ExAC gnomAD |
|
| rs958214404 | 626 | E>K | No | Ensembl | |
| rs373408649 | 628 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs199679586 | 629 | R>L | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs199679586 | 629 | R>Q | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs2071378556 | 630 | L>R | No | Ensembl | |
| rs2071378273 | 630 | L>V | No | TOPMed | |
| rs2071378901 | 631 | K>N | No |
TOPMed gnomAD |
|
| rs776341807 | 631 | K>T | No | ExAC | |
| TCGA novel | 634 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2071379449 | 635 | Q>E | No | Ensembl | |
| rs2071379804 | 636 | E>G | No | Ensembl | |
| rs1248248548 | 637 | R>Q | No |
TOPMed gnomAD |
|
| rs1468362344 | 639 | I>M | No |
TOPMed gnomAD |
|
| rs761478455 | 640 | I>T | No |
ExAC gnomAD |
|
| COSM1307755 | 643 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1602272646 | 648 | E>A | No | Ensembl | |
| rs867011198 | 649 | A>D | No | Ensembl | |
| rs1160518695 | 650 | Q>H | No | gnomAD | |
|
COSM4912705 COSM4912704 COSM4912706 |
651 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749877076 | 651 | R>K | No |
ExAC gnomAD |
|
| rs762631985 | 652 | R>L | No |
ExAC gnomAD |
|
| rs762631985 | 652 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2071645057 | 653 | A>V | No | Ensembl | |
|
rs2148042910 RCV001579317 |
654 | Q>* | No |
ClinVar Ensembl dbSNP |
|
| rs765849633 | 654 | Q>P | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 655 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 657 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1326003876 | 657 | D>H | No | TOPMed | |
| rs374968601 | 658 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs180730335 | 658 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2071646735 | 659 | Q>L | No | TOPMed | |
| rs757686884 | 660 | W>R | No |
ExAC gnomAD |
|
| rs2071647119 | 661 | L>P | No | TOPMed | |
| rs779489217 | 662 | E>G | No |
ExAC gnomAD |
|
| rs750646540 | 663 | H>D | No |
ExAC gnomAD |
|
| rs758744871 | 663 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs750646540 | 663 | H>Y | No |
ExAC gnomAD |
|
| rs1241486801 | 665 | Q>H | No | gnomAD | |
|
COSM5130514 COSM1413912 COSM5130513 |
666 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1460959741 | 666 | Q>P | No |
TOPMed gnomAD |
|
| rs1460959741 | 666 | Q>R | No |
TOPMed gnomAD |
|
| rs2148043473 | 668 | D>G | No | Ensembl | |
| rs1188308312 | 668 | D>N | No | gnomAD | |
|
COSM4295841 COSM4295840 COSM2844113 |
669 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1385104755 | 673 | P>L | No | gnomAD | |
| rs747091824 | 674 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1569199924 | 675 | K>R | No | TOPMed | |
|
rs376139396 COSM3550437 COSM3550436 COSM3550438 |
678 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs201545935 | 679 | E>K | No | gnomAD | |
| rs201545935 | 679 | E>Q | No | gnomAD | |
| rs1569199991 | 679 | E>V | No | Ensembl | |
| rs2071650993 | 681 | K>N | No | Ensembl | |
| COSM1030408 | 681 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2071651372 | 683 | K>E | No | gnomAD | |
| rs769546913 | 685 | E>K | No |
ExAC gnomAD |
|
| rs769546913 | 685 | E>Q | No |
ExAC gnomAD |
|
| rs762540696 | 687 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1429953136 | 687 | S>R | No | gnomAD | |
| rs2071652550 | 689 | K>E | No |
TOPMed gnomAD |
|
| rs1342197848 | 689 | K>R | No |
TOPMed gnomAD |
|
| rs1278708747 | 690 | K>T | No | gnomAD | |
| rs139761574 | 691 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs1221435018 | 692 | D>G | No |
TOPMed gnomAD |
|
| rs1000332106 | 692 | D>N | No | TOPMed | |
| rs758978661 | 694 | E>K | No |
ExAC TOPMed gnomAD |
|
|
rs972530973 COSM220441 |
695 | E>G | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated Ensembl |
|
COSM4746961 COSM4746960 COSM4746959 |
697 | G>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1201177706 | 701 | A>T | No | gnomAD | |
| rs1412252919 | 702 | Q>H | No | TOPMed | |
| rs1322102424 | 702 | Q>K | No |
TOPMed gnomAD |
|
| rs1280835920 | 704 | K>R | No | gnomAD | |
| rs762453980 | 707 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754299315 | 707 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2148044641 | 708 | L>H | No | Ensembl | |
| rs141317839 | 708 | L>V | No |
1000Genomes gnomAD |
|
| rs1569200437 | 709 | F>L | No | Ensembl | |
| rs2071657641 | 709 | F>L | No | TOPMed | |
| rs1569200437 | 709 | F>V | No | Ensembl | |
| rs2071658260 | 710 | H>R | No | Ensembl | |
| rs2071658005 | 710 | H>Y | No | Ensembl | |
| rs1336899139 | 712 | H>R | No | gnomAD | |
| rs2071659039 | 715 | P>R | No | gnomAD | |
| rs1602292961 | 715 | P>S | No | Ensembl | |
| rs1479033943 | 716 | A>V | No | gnomAD | |
| rs2071659631 | 717 | K>E | No | Ensembl | |
| rs765678311 | 720 | V>I | No |
ExAC gnomAD |
|
| rs2071660016 | 722 | A>T | No | Ensembl | |
| rs2071660464 | 724 | W>* | No | Ensembl | |
| rs1243067624 | 724 | W>C | No | gnomAD | |
| rs2071660896 | 725 | S>C | No | Ensembl | |
|
COSM4101406 COSM4101408 COSM4101407 |
727 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1382167932 | 728 | E>K | No | gnomAD | |
| rs1336515977 | 729 | K>R | No |
TOPMed gnomAD |
|
| rs2071841909 | 730 | G>D | No | TOPMed | |
| rs2071841909 | 730 | G>V | No | TOPMed | |
| rs763242335 | 731 | P>A | No |
ExAC gnomAD |
|
| rs145376740 | 737 | Q>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs950873983 | 738 | E>D | No | TOPMed | |
| rs751698164 | 738 | E>K | No |
ExAC gnomAD |
|
| rs2071844112 | 741 | K>Q | No | TOPMed | |
| rs2071844808 | 743 | V>E | No | TOPMed | |
| rs755276554 | 743 | V>M | No |
ExAC gnomAD |
|
| rs985361819 | 746 | R>Q | No |
TOPMed gnomAD |
|
| rs374245664 | 746 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1159854030 | 747 | A>S | No | TOPMed | |
|
COSM4937069 COSM4937070 COSM4937071 |
747 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2071846306 | 748 | L>V | No | TOPMed | |
| rs1602306998 | 749 | Y>S | No | Ensembl | |
| rs2071846905 | 750 | P>L | No |
TOPMed gnomAD |
|
| rs2071846905 | 750 | P>R | No |
TOPMed gnomAD |
|
| rs1231890672 | 754 | R>G | No |
TOPMed gnomAD |
|
| rs2071847376 | 755 | S>G | No | gnomAD | |
| rs1442190994 | 755 | S>R | No | gnomAD | |
| rs1162891760 | 756 | H>R | No | gnomAD | |
| TCGA novel | 759 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1030409 | 760 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2071847985 | 760 | T>S | No | Ensembl | |
| rs909416894 | 761 | I>V | No |
TOPMed gnomAD |
|
| rs2071848418 | 762 | Q>* | No | TOPMed | |
| rs2071848932 | 762 | Q>H | No |
TOPMed gnomAD |
|
| rs2071848735 | 762 | Q>R | No | TOPMed | |
| rs201107589 | 763 | P>A | No |
1000Genomes TOPMed |
|
| rs201107589 | 763 | P>S | No |
1000Genomes TOPMed |
|
| rs755974128 | 765 | D>N | No |
ExAC gnomAD |
|
|
COSM4693650 rs2071849815 COSM4693652 COSM4693651 |
768 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs774801131 | 770 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1402702053 | 771 | G>R | No | gnomAD | |
| TCGA novel | 772 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs898961805 | 772 | E>K | No |
TOPMed gnomAD |
|
| COSM2150806 | 773 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 774 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2072870438 | 777 | S>I | No | TOPMed | |
| rs1426099480 | 780 | G>A | No | gnomAD | |
| rs1400820234 | 780 | G>R | No | TOPMed | |
| rs1035683296 | 781 | E>Q | No | Ensembl | |
| rs2148187663 | 782 | P>S | No | Ensembl | |
| rs1156923746 | 783 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes NCI-TCGA TOPMed gnomAD |
| rs2072871584 | 783 | G>D | No | TOPMed | |
| rs1156923746 | 783 | G>S | No |
1000Genomes TOPMed gnomAD |
|
| rs2148187766 | 784 | W>* | No | Ensembl | |
|
COSM6094704 COSM6094705 COSM6094706 |
787 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3841835 COSM3841837 COSM3841836 |
790 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760929885 | 791 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs376275091 | 793 | T>I | No |
ESP TOPMed gnomAD |
|
| rs2072872359 | 794 | G>R | No | Ensembl | |
| rs2072872359 | 794 | G>W | No | Ensembl | |
|
COSM1307756 rs2148187943 |
795 | W>* | urinary_tract Variant assessed as Somatic; HIGH impact. [Cosmic, NCI-TCGA] | No |
cosmic curated Ensembl NCI-TCGA Cosmic |
| rs2148188031 | 798 | A>T | No | Ensembl | |
| rs2072872875 | 798 | A>V | No | Ensembl | |
|
COSM1751620 rs2072873268 COSM3933947 |
800 | Y>C | urinary_tract [Cosmic] | No |
cosmic curated Ensembl |
| rs267606114 | 800 | Y>N | No | Ensembl | |
| rs1364132282 | 801 | A>T | No | gnomAD | |
| rs2148188162 | 801 | A>V | No | Ensembl | |
| rs2072873952 | 802 | E>D | No | Ensembl | |
| rs1436168446 | 803 | K>E | No | gnomAD | |
| rs753977647 | 804 | I>N | No |
ExAC gnomAD |
|
|
COSM3550441 COSM3550439 COSM3550440 |
805 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2148188372 | 807 | N>K | No | Ensembl | |
| rs781033642 | 809 | V>D | No | Ensembl | |
| rs2072875202 | 810 | P>R | No | TOPMed | |
| rs2148188482 | 810 | P>S | No | Ensembl | |
| rs201031751 | 811 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780063779 | 812 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1289458348 | 812 | P>L | No | gnomAD | |
| rs2148188670 | 816 | V>E | No | Ensembl | |
| rs751233796 | 816 | V>L | No |
ExAC gnomAD |
|
| rs751233796 | 816 | V>M | No |
ExAC gnomAD |
|
| rs1188810030 | 817 | T>A | No | gnomAD | |
| rs780808746 | 818 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs2072876862 | 818 | D>Y | No | TOPMed | |
| rs2148188822 | 819 | S>L | No | Ensembl | |
| rs747697396 | 820 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs2148188881 | 820 | T>I | No | Ensembl | |
| rs771280043 | 822 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs771280043 | 822 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs779459072 | 823 | P>R | No |
ExAC gnomAD |
|
| rs2072878000 | 823 | P>S | No | Ensembl | |
| rs746425456 | 824 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs772397163 | 825 | P>H | No |
ExAC gnomAD |
|
|
COSM3912045 COSM3912046 COSM3912044 |
825 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1428323420 | 826 | K>Q | No | gnomAD | |
| rs1351914489 | 826 | K>R | No | gnomAD | |
| rs775884476 | 827 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2148189361 | 828 | A>D | No | Ensembl | |
| rs760841811 | 828 | A>T | No |
ExAC gnomAD |
|
|
COSM5366317 COSM5366316 COSM5366318 |
828 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs768901601 COSM725235 COSM5131672 COSM5131671 |
830 | R>C | lung Variant assessed as Somatic; MODERATE impact. large_intestine [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs1028088112 COSM4101410 COSM4101409 COSM4101411 |
830 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1459798087 | 831 | E>G | No | gnomAD | |
| rs776509377 | 832 | T>A | No | ExAC | |
| rs776509377 | 832 | T>P | No | ExAC | |
| rs776509377 | 832 | T>S | No | ExAC | |
| rs2148189577 | 833 | P>H | No | Ensembl | |
| rs2072880837 | 833 | P>S | No | TOPMed | |
| rs2072880837 | 833 | P>T | No | TOPMed | |
| rs765293456 | 834 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs765293456 | 834 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM444400 rs765293456 COSM5227680 COSM5227679 |
834 | A>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 834 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1228743977 | 835 | P>L | No | gnomAD | |
| rs2148189826 | 836 | L>V | No | Ensembl | |
| rs908244701 | 837 | A>T | No | gnomAD | |
| rs2148189896 | 837 | A>V | No | Ensembl | |
| rs2148189924 | 838 | V>I | No | Ensembl | |
|
COSM4942083 COSM4942082 COSM4942081 |
838 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs369327593 | 839 | T>I | No |
ESP TOPMed |
|
| rs369327593 | 839 | T>N | No |
ESP TOPMed |
|
| rs1602378590 | 840 | S>P | No | Ensembl | |
| rs2148190083 | 843 | P>S | No | Ensembl | |
| rs1180573036 | 844 | S>F | No | gnomAD | |
| rs200510636 | 845 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1602378692 | 845 | T>P | No | Ensembl | |
| rs766225849 | 846 | T>P | No |
ExAC gnomAD |
|
| rs1260485260 | 847 | P>R | No | TOPMed | |
| rs754610672 | 848 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs754610672 | 848 | N>T | No |
ExAC TOPMed gnomAD |
|
|
COSM3841840 COSM3841839 rs2072885449 COSM3841838 |
851 | A>T | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs780907492 | 851 | A>V | No |
ExAC gnomAD |
|
| rs2072886212 | 852 | D>G | No | TOPMed | |
| rs567513630 | 852 | D>N | No |
1000Genomes ExAC gnomAD |
|
| rs1602379048 | 853 | F>L | No | Ensembl | |
| rs1339309675 | 854 | S>G | No | gnomAD | |
| rs2072887152 | 855 | S>C | No |
TOPMed gnomAD |
|
| rs746355159 | 856 | T>A | No |
ExAC gnomAD |
|
| rs772592802 | 856 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs746355159 | 856 | T>P | No |
ExAC gnomAD |
|
| rs2073093473 | 857 | W>C | No | Ensembl | |
| rs1264371857 | 858 | P>S | No |
1000Genomes TOPMed gnomAD |
|
| rs781452040 | 861 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs781452040 | 861 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs1478102347 | 862 | N>D | No | gnomAD | |
| rs141514423 | 863 | E>G | No |
ESP TOPMed gnomAD |
|
| rs2073094623 | 863 | E>K | No | TOPMed | |
| rs1173814511 | 864 | K>E | No | gnomAD | |
| rs1419033938 | 866 | E>G | No | gnomAD | |
| rs1377875641 | 866 | E>Q | No |
TOPMed gnomAD |
|
| rs377643560 | 867 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs377643560 | 867 | T>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1569234672 | 868 | D>E | No | Ensembl | |
| rs2148218587 | 871 | D>Y | No | Ensembl | |
| rs774422281 | 872 | A>G | No |
ExAC gnomAD |
|
| rs1167808813 | 873 | W>R | No | TOPMed | |
| rs775173579 | 874 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs2073096686 | 874 | A>T | No | TOPMed | |
| rs775173579 | 874 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs199689124 | 875 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs768966402 | 875 | A>V | No | Ensembl | |
| rs924585580 | 877 | P>A | No | TOPMed | |
| rs1305729052 | 877 | P>R | No | gnomAD | |
| rs1351533048 | 878 | S>C | No |
TOPMed gnomAD |
|
| rs1351533048 | 878 | S>F | No |
TOPMed gnomAD |
|
| rs756747937 | 880 | T>A | No |
ExAC gnomAD |
|
| rs1266314621 | 881 | V>A | No | gnomAD | |
| rs147036952 | 881 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1569235001 | 883 | S>G | No | Ensembl | |
| rs373166988 | 885 | G>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373166988 | 885 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs140905223 | 885 | G>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2073100019 | 886 | Q>L | No | Ensembl | |
| rs1438500944 | 890 | R>K | No | gnomAD | |
| rs1602395126 | 891 | S>A | No | Ensembl | |
| rs778127066 | 892 | A>S | No |
ExAC gnomAD |
|
| rs778127066 | 892 | A>T | No |
ExAC gnomAD |
|
| rs2073101116 | 895 | P>L | No | TOPMed | |
| rs2073101270 | 896 | A>P | No | Ensembl | |
| rs933553358 | 897 | T>A | No | Ensembl | |
|
COSM1030411 rs377099843 |
897 | T>M | endometrium [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs779181323 | 899 | T>A | No |
ExAC gnomAD |
|
| rs2073102513 | 900 | G>S | No | gnomAD | |
| rs1424758584 | 902 | S>C | No |
TOPMed gnomAD |
|
| rs1602395360 | 902 | S>P | No | Ensembl | |
| rs143723211 | 903 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs745772242 | 903 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs745772242 | 903 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1375428962 | 904 | S>C | No | TOPMed | |
|
COSM4398111 COSM4398110 COSM4398112 |
905 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1371769254 | 910 | G>D | No |
TOPMed gnomAD |
|
| rs1434301162 | 912 | K>E | No | TOPMed | |
| rs1331564845 | 913 | V>M | No | gnomAD | |
| rs758698817 | 915 | G>W | No |
ExAC gnomAD |
|
| rs751693562 | 919 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2073421848 | 923 | P>S | No | Ensembl | |
| rs900882628 | 926 | A>G | No | Ensembl | |
| rs781090343 | 926 | A>S | No |
ExAC gnomAD |
|
| rs781090343 | 926 | A>T | No |
ExAC gnomAD |
|
| rs2073422593 | 928 | K>E | No | TOPMed | |
| rs2073422913 | 929 | D>G | No | TOPMed | |
| rs1185383739 | 929 | D>N | No | gnomAD | |
| TCGA novel | 929 | D>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1216843108 | 930 | N>D | No |
TOPMed gnomAD |
|
| rs1216843108 | 930 | N>H | No |
TOPMed gnomAD |
|
| rs1049483014 | 930 | N>S | No | TOPMed | |
| rs1441912954 | 931 | H>R | No |
TOPMed gnomAD |
|
| rs1259845651 | 933 | N>H | No | gnomAD | |
| rs2073424385 | 935 | N>K | No | TOPMed | |
| rs1487452457 | 935 | N>S | No | gnomAD | |
| rs755867780 | 936 | K>R | No |
ExAC gnomAD |
|
| rs777671752 | 938 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs777671752 | 938 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs777671752 | 938 | D>Y | No |
ExAC TOPMed gnomAD |
|
| rs748843237 | 939 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1170741059 | 940 | I>V | No | gnomAD | |
| rs770646771 | 941 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs773666205 | 941 | T>I | No |
ExAC gnomAD |
|
| rs370969706 | 942 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs370969706 | 942 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1569244104 | 944 | E>Q | No | Ensembl | |
| rs777273587 | 946 | Q>E | No | ExAC | |
| rs762454074 | 948 | M>L | No |
ExAC gnomAD |
|
| rs762454074 | 948 | M>V | No |
ExAC gnomAD |
|
| rs1569244177 | 949 | W>* | No | Ensembl | |
| rs1025382610 | 951 | F>S | No | TOPMed | |
| rs765501018 | 952 | G>A | No |
ExAC gnomAD |
|
| rs1944160043 | 953 | E>G | No | TOPMed | |
| rs763048674 | 954 | V>F | No |
ExAC gnomAD |
|
| rs763048674 | 954 | V>I | No |
ExAC gnomAD |
|
| rs2148264599 | 955 | Q>H | No | Ensembl | |
| rs766634928 | 956 | G>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 957 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1311575417 | 957 | Q>R | No |
TOPMed gnomAD |
|
| rs751833996 | 958 | K>N | No |
ExAC gnomAD |
|
| rs2073429083 | 961 | F>C | No | Ensembl | |
| rs2073428921 | 961 | F>V | No | Ensembl | |
| rs759604948 | 963 | K>R | No |
ExAC gnomAD |
|
|
RCV001579318 rs2148264796 |
965 | Y>* | No |
ClinVar Ensembl dbSNP |
|
| rs2073429744 | 966 | V>L | No | TOPMed | |
| rs2073429744 | 966 | V>M | No | TOPMed | |
| rs1216174168 | 971 | G>A | No | gnomAD | |
| rs1216174168 | 971 | G>E | No | gnomAD | |
|
COSM6094700 rs1448509034 COSM578890 COSM6094699 |
973 | I>V | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM4101413 COSM4101412 |
975 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1246027463 | 976 | S>F | No | gnomAD | |
| rs539887450 | 977 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs558186603 | 977 | T>I | No |
1000Genomes ExAC gnomAD |
|
| COSM1632467 | 978 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1460213279 | 980 | D>H | No |
TOPMed gnomAD |
|
| rs2073492189 | 984 | S>L | No | Ensembl | |
| rs367925531 | 986 | S>G | No |
ESP TOPMed gnomAD |
|
| rs758150653 | 986 | S>N | No |
ExAC gnomAD |
|
| rs1428479472 | 988 | A>G | No | TOPMed | |
| rs2073493263 | 989 | S>I | No | TOPMed | |
| rs1213077839 | 989 | S>R | No | gnomAD | |
| rs372156010 | 990 | L>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1193011407 | 990 | L>P | No |
TOPMed gnomAD |
|
| rs372156010 | 990 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM6008590 COSM1177139 rs1242902737 |
992 | R>* | Variant assessed as Somatic; HIGH impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs200116329 | 992 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2073493967 | 993 | V>A | No | TOPMed | |
| rs200850499 | 994 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs754456971 | 995 | S>C | No | Ensembl | |
| rs2073494825 | 996 | P>L | No | TOPMed | |
| rs778336685 | 996 | P>S | No |
ExAC gnomAD |
|
| rs1256054576 | 998 | A>G | No |
TOPMed gnomAD |
|
| rs2073495916 | 999 | K>N | No | Ensembl | |
| rs749796773 | 1000 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs771300835 | 1001 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs774720789 | 1002 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1475729176 | 1003 | S>A | No | gnomAD | |
|
COSM5629129 rs772284990 COSM222410 |
1003 | S>L | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs764148741 | 1004 | G>R | No |
ExAC gnomAD |
|
| rs199861309 | 1006 | E>A | No |
1000Genomes TOPMed |
|
| rs199861309 | 1006 | E>G | No |
1000Genomes TOPMed |
|
| rs2148308597 | 1007 | F>C | No | Ensembl | |
| TCGA novel | 1007 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2073808547 | 1008 | I>M | No | Ensembl | |
| TCGA novel | 1009 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765273903 | 1010 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs761774416 | 1010 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs761774416 | 1010 | M>R | No |
ExAC TOPMed gnomAD |
|
| rs761774416 | 1010 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs2073808729 | 1010 | M>V | No | TOPMed | |
| rs773086924 | 1011 | Y>N | No |
ExAC gnomAD |
|
| rs2073809682 | 1012 | T>A | No | TOPMed | |
| rs2073809855 | 1013 | Y>* | No | TOPMed | |
|
COSM5829099 COSM1413913 |
1013 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1485739200 | 1014 | E>K | No | gnomAD | |
| rs2073810404 | 1015 | S>N | No | Ensembl | |
| rs751198160 | 1017 | E>K | No |
ExAC gnomAD |
|
| rs2073810720 | 1023 | F>V | No | TOPMed | |
|
COSM4101415 COSM4101414 |
1025 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1243348010 | 1027 | D>A | No | TOPMed | |
| rs935811731 | 1028 | V>M | No | TOPMed | |
| rs922048723 | 1033 | K>N | No | TOPMed | |
| rs757702456 | 1034 | K>E | No |
ExAC gnomAD |
|
|
COSM312096 rs2148309198 RCV001579324 |
1039 | W>* | lung [Cosmic] | No |
cosmic curated ClinVar Ensembl dbSNP |
| rs932040702 | 1042 | T>R | No |
TOPMed gnomAD |
|
| rs746443067 | 1044 | G>R | No |
ExAC gnomAD |
|
| rs534329964 | 1045 | D>E | No | Ensembl | |
| rs1439178942 | 1045 | D>N | No |
TOPMed gnomAD |
|
| rs758734169 | 1047 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs2148309440 | 1047 | A>V | No | Ensembl | |
|
COSM5440786 COSM5440785 rs1440104219 |
1048 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs747205867 | 1049 | V>A | No |
ExAC gnomAD |
|
| TCGA novel | 1050 | F>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1569254945 | 1052 | S>C | No | Ensembl | |
|
COSM4101417 COSM4101416 |
1052 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1283093180 | 1054 | Y>C | No | gnomAD | |
| rs890418146 | 1060 | S>A | No |
TOPMed gnomAD |
|
| TCGA novel | 1061 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs571982570 | 1062 | G>S | No |
1000Genomes ExAC gnomAD |
|
| rs778018287 | 1064 | G>E | No |
ExAC gnomAD |
|
| rs1359667414 | 1065 | T>S | No |
TOPMed gnomAD |
|
| rs1281209283 | 1065 | T>S | No | gnomAD | |
| rs2074000724 | 1066 | A>S | No | Ensembl | |
| rs749194076 | 1067 | G>E | No |
ExAC gnomAD |
|
| rs770996681 | 1069 | T>A | No |
ExAC gnomAD |
|
| rs774137935 | 1070 | G>V | No |
ExAC gnomAD |
|
| rs1419889472 | 1071 | S>R | No | gnomAD | |
| rs1315885012 | 1072 | L>S | No |
TOPMed gnomAD |
|
| rs759236714 | 1073 | G>R | No |
ExAC gnomAD |
|
|
COSM1641456 COSM4336863 |
1075 | K>N | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2074002104 | 1075 | K>N | No | TOPMed | |
| rs2074002242 | 1076 | P>L | No | TOPMed | |
| TCGA novel | 1076 | P>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs961259059 | 1077 | E>D | No | Ensembl | |
| rs2074175871 | 1080 | Q>R | No | TOPMed | |
| rs771743810 | 1081 | V>A | No |
ExAC gnomAD |
|
| rs771743810 | 1081 | V>D | No |
ExAC gnomAD |
|
| rs1569267041 | 1081 | V>I | No | Ensembl | |
| rs200073587 | 1082 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1448347335 | 1083 | A>T | No | Ensembl | |
| TCGA novel | 1085 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs936698706 | 1086 | T>A | No | TOPMed | |
| rs1297739490 | 1086 | T>I | No | gnomAD | |
|
COSM1413914 rs913872456 |
1087 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA gnomAD |
| rs767729155 | 1089 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs760108413 | 1089 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs760108413 | 1089 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs767729155 | 1089 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1325641330 | 1090 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
COSM4819535 COSM4819536 rs2074179091 |
1091 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1453844010 | 1095 | L>V | No |
TOPMed gnomAD |
|
| rs764397454 | 1096 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1156420285 | 1096 | A>V | No | TOPMed | |
| rs754177477 | 1097 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs754177477 | 1097 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1272439562 | 1098 | G>A | No |
TOPMed gnomAD |
|
| COSM1030415 | 1098 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1188656204 | 1099 | Q>L | No |
TOPMed gnomAD |
|
| rs535264292 | 1104 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1211185 rs535264292 |
1104 | R>Q | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs745745415 | 1105 | K>E | No |
ExAC gnomAD |
|
| rs554932669 | 1105 | K>I | No |
ExAC gnomAD |
|
| rs554932669 | 1105 | K>R | No |
ExAC gnomAD |
|
| rs1365241774 | 1108 | P>R | No | gnomAD | |
| rs1569267628 | 1109 | G>C | No | Ensembl | |
| rs1163272581 | 1109 | G>D | No | gnomAD | |
| rs1417721771 | 1110 | G>* | No | gnomAD | |
| rs201077384 | 1112 | W>C | No | 1000Genomes | |
| rs768502491 | 1114 | G>E | No |
ExAC gnomAD |
|
| rs1295300557 | 1115 | E>K | No | gnomAD | |
| rs758212573 | 1118 | A>T | No |
ExAC gnomAD |
|
| rs1235101198 | 1118 | A>V | No | gnomAD | |
| rs779957035 | 1119 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs2074475719 | 1119 | R>H | No | TOPMed | |
| rs747021058 | 1122 | K>Q | No |
ExAC gnomAD |
|
| rs1325821849 | 1123 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs754740442 | 1124 | Q>R | No |
ExAC TOPMed gnomAD |
|
|
COSM3405373 rs2074476278 COSM3405374 |
1126 | G>S | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs2074476418 | 1126 | G>V | No | TOPMed | |
|
COSM3550448 COSM3550447 |
1129 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs199990852 | 1130 | A>S | No |
TOPMed gnomAD |
|
| rs2074476662 | 1130 | A>V | No |
TOPMed gnomAD |
|
| rs780983949 | 1132 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs780983949 | 1132 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs780983949 | 1132 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs1209704617 | 1133 | V>I | No |
TOPMed gnomAD |
|
| rs747710795 | 1134 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs769489097 | 1135 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs777086928 | 1136 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs2074477819 | 1137 | S>G | No | TOPMed | |
|
VAR_070011 rs187895245 |
1137 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
UniProt 1000Genomes ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs201419866 | 1138 | P>R | No |
1000Genomes ExAC gnomAD |
|
| rs200762568 | 1140 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1333101773 | 1141 | S>G | No | TOPMed | |
| rs534506275 | 1141 | S>N | No | 1000Genomes | |
| rs1392108558 | 1142 | K>T | No |
TOPMed gnomAD |
|
| rs1004780994 | 1143 | I>M | No | TOPMed | |
| rs1385984924 | 1143 | I>V | No |
TOPMed gnomAD |
|
| rs761313234 | 1146 | T>A | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1148 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769093517 | 1151 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1384093143 | 1152 | T>S | No | gnomAD | |
| rs777085854 | 1153 | A>V | No |
ExAC gnomAD |
|
| rs200818653 | 1155 | A>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200818653 | 1155 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs750672753 | 1156 | A>E | No |
ExAC gnomAD |
|
| rs755904968 | 1157 | V>A | No |
ExAC gnomAD |
|
| rs777478671 | 1158 | C>S | No |
ExAC gnomAD |
|
| rs753481055 | 1160 | V>A | No |
ExAC gnomAD |
|
| rs267606115 | 1162 | G>R | No | Ensembl | |
| rs201693658 | 1165 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs745386929 | 1167 | T>I | No |
ExAC gnomAD |
|
| rs185723649 | 1168 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs748645246 | 1168 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2074610995 | 1169 | Q>* | No | Ensembl | |
| rs773476019 | 1170 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1471893861 | 1172 | D>N | No | TOPMed | |
| rs139755461 | 1176 | F>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs188915920 | 1177 | N>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs767532845 | 1177 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs767532845 | 1177 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs760552809 | 1179 | G>D | No |
ExAC gnomAD |
|
| rs2074612609 | 1180 | Q>R | No | Ensembl | |
| rs2074612859 | 1181 | I>T | No | Ensembl | |
| rs763908413 | 1181 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs372383366 | 1183 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs764971322 | 1184 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs764971322 | 1184 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1169168864 | 1185 | L>F | No | gnomAD | |
| rs1370734182 | 1185 | L>H | No | gnomAD | |
| TCGA novel | 1187 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779481974 | 1187 | K>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1188 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1602536185 | 1190 | P>L | No |
TOPMed gnomAD |
|
| rs1310168098 | 1190 | P>S | No | gnomAD | |
| rs756580360 | 1191 | D>A | No |
ExAC gnomAD |
|
| TCGA novel | 1191 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2074614603 | 1191 | D>Y | No | TOPMed | |
| rs2074614869 | 1192 | W>C | No | gnomAD | |
| rs778136789 | 1193 | W>R | No | ExAC | |
| rs2074615127 | 1194 | K>R | No |
TOPMed gnomAD |
|
| TCGA novel | 1195 | G>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM115686 | 1195 | G>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs376413318 | 1198 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1202 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM725234 rs1382008003 |
1206 | S>F | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1207 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs929342450 | 1207 | N>S | No | TOPMed | |
| rs984751447 | 1208 | Y>C | No |
TOPMed gnomAD |
|
| rs984751447 | 1208 | Y>F | No |
TOPMed gnomAD |
|
| rs1343061780 | 1212 | T>I | No | gnomAD | |
| rs1278532338 | 1213 | T>A | No | gnomAD | |
|
COSM6094697 COSM6094698 |
1214 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2074616934 | 1215 | M>R | No | gnomAD | |
| rs2074616934 | 1215 | M>T | No | gnomAD | |
| rs772141065 | 1215 | M>V | No |
ExAC gnomAD |
|
| rs1602536527 | 1216 | D>A | No | TOPMed | |
| rs1602536527 | 1216 | D>G | No | TOPMed | |
| rs1602536577 | 1217 | P>Q | No | Ensembl | |
| rs374844830 | 1219 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs2074617867 | 1220 | Q>H | No | TOPMed | |
| rs760594941 | 1220 | Q>P | No |
ExAC gnomAD |
|
| rs2074617994 | 1221 | W>G | No | TOPMed | |
|
COSM6094694 COSM6094693 |
1221 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1979414884 | 1224 | D>E | No |
TOPMed gnomAD |
|
|
COSM3550450 COSM3550449 |
1224 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1225 | L>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs527375628 | 1226 | H>L | No |
1000Genomes ExAC gnomAD |
|
| rs527375628 | 1226 | H>R | No |
1000Genomes ExAC gnomAD |
|
| rs1303504348 | 1226 | H>Y | No | TOPMed | |
| rs1979416947 | 1228 | L>S | No | gnomAD | |
| rs1979417895 | 1230 | M>V | No | gnomAD | |
| rs1158929382 | 1232 | T>I | No | gnomAD | |
| rs542782139 | 1234 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1275928791 | 1234 | T>I | No |
TOPMed gnomAD |
|
| rs1275928791 | 1234 | T>N | No |
TOPMed gnomAD |
|
| rs769863378 | 1236 | R>G | No |
ExAC gnomAD |
|
| rs1162793857 | 1237 | K>R | No | gnomAD | |
| rs997210138 | 1238 | R>* | No | Ensembl | |
| rs201178787 | 1238 | R>Q | No |
TOPMed gnomAD |
|
| rs773336718 | 1241 | Y>H | No |
ExAC gnomAD |
|
| rs770796256 | 1243 | H>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1243 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM2844168 rs1979424956 |
1244 | E>K | liver [Cosmic] | No |
cosmic curated Ensembl |
| rs1979425412 | 1245 | L>F | No | 1000Genomes | |
| rs1979425412 | 1245 | L>I | No | 1000Genomes | |
| rs1331045533 | 1246 | I>L | No | gnomAD | |
| rs1231375373 | 1246 | I>T | No |
TOPMed gnomAD |
|
| rs1264857466 | 1247 | V>I | No | gnomAD | |
|
COSM5057573 COSM1030417 rs1211156247 |
1249 | E>K | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| TCGA novel | 1250 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1028461442 | 1250 | E>K | No |
TOPMed gnomAD |
|
| rs1181836739 | 1253 | V>M | No | gnomAD | |
| TCGA novel | 1253 | V>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1255 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752206485 | 1256 | L>V | No |
ExAC gnomAD |
|
| rs1379242671 | 1258 | L>V | No | TOPMed | |
| rs762248759 | 1259 | V>F | No |
ExAC gnomAD |
|
| rs1979856981 | 1262 | I>V | No | gnomAD | |
| COSM261472 | 1264 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1246781777 | 1268 | M>I | No | gnomAD | |
| rs1979859450 | 1269 | E>A | No | Ensembl | |
| rs967427079 | 1269 | E>D | No | Ensembl | |
| rs2148482777 | 1269 | E>K | No | Ensembl | |
| rs2148482799 | 1271 | E>K | No | Ensembl | |
| rs751961174 | 1274 | T>A | No |
ExAC gnomAD |
|
| rs1346918496 | 1275 | E>G | No |
TOPMed gnomAD |
|
| rs1979861530 | 1277 | E>Q | No | gnomAD | |
| TCGA novel | 1277 | E>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs149830687 | 1278 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1393077084 | 1279 | A>S | No |
TOPMed gnomAD |
|
| rs1392206544 | 1280 | M>L | No | gnomAD | |
| rs1569310168 | 1280 | M>R | No | Ensembl | |
| rs767843094 | 1282 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 1282 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs753004103 | 1283 | V>M | No |
ExAC gnomAD |
|
|
COSM3405377 COSM3405378 |
1284 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1431331665 | 1290 | M>I | No | gnomAD | |
| rs1979866495 | 1290 | M>V | No | Ensembl | |
| rs374237907 | 1292 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV002267343 rs2148501075 |
1298 | A>V | No |
ClinVar Ensembl dbSNP |
|
| rs1418779509 | 1300 | R>K | No | TOPMed | |
|
COSM5097436 COSM1413915 |
1302 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746627172 | 1304 | K>M | No |
ExAC TOPMed gnomAD |
|
| rs1981340670 | 1306 | S>C | No | Ensembl | |
| rs1179675954 | 1307 | G>R | No | gnomAD | |
| rs1445959499 | 1308 | E>D | No |
TOPMed gnomAD |
|
| TCGA novel | 1308 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1981343099 | 1308 | E>K | No | Ensembl | |
| rs1981344499 | 1309 | K>R | No | TOPMed | |
| rs1472829297 | 1310 | M>I | No | gnomAD | |
| rs1259051665 | 1310 | M>V | No |
TOPMed gnomAD |
|
| rs1184231848 | 1311 | P>S | No | gnomAD | |
| rs780514538 | 1312 | V>L | No |
ExAC gnomAD |
|
| rs747685000 | 1313 | K>R | No |
ExAC gnomAD |
|
| rs950936844 | 1314 | M>I | No | Ensembl | |
| rs769223637 | 1315 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs774973776 | 1318 | I>S | No |
ExAC gnomAD |
|
| rs199694383 | 1321 | A>S | No |
1000Genomes ExAC gnomAD |
|
| rs199694383 | 1321 | A>T | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 1322 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1410877412 | 1324 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1377661948 | 1326 | M>L | No | gnomAD | |
| rs965833103 | 1330 | I>V | No | TOPMed | |
|
COSM5461018 COSM5461017 |
1331 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2148501357 | 1331 | R>L | No | Ensembl | |
| rs1272467370 | 1333 | C>R | No | Ensembl | |
| rs1284036421 | 1334 | S>R | No |
TOPMed gnomAD |
|
| rs2148501389 | 1334 | S>T | No | Ensembl | |
| rs201465943 | 1335 | R>C | No |
ExAC gnomAD |
|
| rs1223546579 | 1335 | R>L | No |
TOPMed gnomAD |
|
|
COSM3550452 COSM3550451 |
1335 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764356694 | 1337 | L>H | No |
ExAC gnomAD |
|
| rs145345122 | 1338 | N>S | No |
ESP ExAC gnomAD |
|
| rs1235619622 | 1339 | G>R | No |
TOPMed gnomAD |
|
| rs147679731 | 1340 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1251151046 | 1341 | A>S | No | gnomAD | |
| rs1251151046 | 1341 | A>T | No | gnomAD | |
| rs779781593 | 1346 | K>N | No |
ExAC TOPMed gnomAD |
|
|
COSM5067650 COSM1566080 |
1346 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781233789 | 1347 | T>K | No | gnomAD | |
|
COSM4752653 rs781233789 COSM4752654 |
1347 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs9976801 | 1348 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1331315779 | 1348 | D>N | No | gnomAD | |
| rs1303993322 | 1349 | E>G | No | gnomAD | |
| rs1447947972 | 1349 | E>K | No |
TOPMed gnomAD |
|
| rs1981366732 | 1351 | P>T | No | TOPMed | |
| rs1024356918 | 1352 | D>E | No | Ensembl | |
| rs1226328098 | 1352 | D>Y | No | gnomAD | |
| rs1981368577 | 1353 | F>I | No |
TOPMed gnomAD |
|
| rs2024913125 | 1354 | K>T | No | TOPMed | |
| rs1230271558 | 1356 | F>L | No | gnomAD | |
|
COSM270936 rs185389697 |
1357 | V>F | large_intestine [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs185389697 | 1357 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776188552 | 1362 | M>I | No | Ensembl | |
| rs762825823 | 1362 | M>V | No |
ExAC gnomAD |
|
| rs1343998353 | 1363 | D>N | No | gnomAD | |
| rs1476149405 | 1364 | P>L | No |
TOPMed gnomAD |
|
| rs1014161765 | 1364 | P>S | No |
TOPMed gnomAD |
|
| rs1014161765 | 1364 | P>T | No |
TOPMed gnomAD |
|
| rs751534711 | 1365 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs754793119 | 1365 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751534711 | 1365 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs56279221 | 1367 | K>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV001693616 rs56279221 |
1367 | K>R | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2148506444 | 1368 | G>A | No | Ensembl | |
| TCGA novel | 1368 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1052822780 | 1371 | L>V | No | gnomAD | |
|
COSM3991923 COSM3991922 |
1372 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780248342 | 1375 | I>M | No | Ensembl | |
| rs1569320677 | 1376 | L>V | No | Ensembl | |
| TCGA novel | 1378 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1477883465 | 1379 | M>I | No | gnomAD | |
| TCGA novel | 1381 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1263461434 | 1381 | R>W | No |
TOPMed gnomAD |
|
| rs1602648581 | 1385 | Y>H | No | Ensembl | |
| rs201262861 | 1387 | L>P | No |
1000Genomes TOPMed gnomAD |
|
| rs2148506549 | 1389 | I>N | No | Ensembl | |
| rs1602648613 | 1390 | K>N | No | Ensembl | |
| rs1207147985 | 1391 | N>S | No | TOPMed | |
| rs1437633267 | 1392 | I>M | No | TOPMed | |
| rs763809003 | 1392 | I>N | No |
ExAC TOPMed gnomAD |
|
| rs1983552260 | 1393 | L>P | No | Ensembl | |
| rs966788642 | 1394 | E>G | No | Ensembl | |
| rs1411172210 | 1395 | N>K | No |
TOPMed gnomAD |
|
| rs1288866521 | 1396 | T>P | No | gnomAD | |
| rs1288866521 | 1396 | T>S | No | gnomAD | |
| rs1983555743 | 1396 | T>S | No | Ensembl | |
| rs1602679325 | 1397 | P>A | No | TOPMed | |
|
COSM3673179 rs1602679325 |
1397 | P>T | prostate [Cosmic] | No |
cosmic curated TOPMed |
| rs753612499 | 1398 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1283096589 | 1398 | E>K | No | gnomAD | |
| rs753612499 | 1398 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1351369998 | 1399 | N>K | No | TOPMed | |
| rs1983558806 | 1399 | N>S | No | TOPMed | |
| rs1482258268 | 1400 | H>Q | No | gnomAD | |
| rs756814425 | 1401 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1286087731 | 1402 | D>E | No | gnomAD | |
| rs778578618 | 1403 | H>P | No |
ExAC gnomAD |
|
| rs1185425306 | 1403 | H>Q | No | gnomAD | |
| rs778578618 | 1403 | H>R | No |
ExAC gnomAD |
|
| rs1910131563 | 1404 | S>G | No | TOPMed | |
| rs747481208 | 1404 | S>N | No |
ExAC gnomAD |
|
| rs1473199365 | 1404 | S>R | No | gnomAD | |
| rs1449481033 | 1407 | K>M | No | TOPMed | |
| rs755469594 | 1408 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs781569279 | 1409 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs781569279 | 1409 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770179954 | 1410 | L>V | No |
ExAC gnomAD |
|
| rs749530031 | 1413 | A>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1415 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs548656402 | 1415 | E>D | No |
ExAC gnomAD |
|
| rs1983568399 | 1417 | C>Y | No | Ensembl | |
|
COSM3963943 rs1294890969 |
1418 | S>C | lung [Cosmic] | No |
cosmic curated gnomAD |
|
COSM6161377 COSM6161378 |
1419 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1983570351 | 1422 | E>D | No | TOPMed | |
|
COSM3912050 rs759471287 COSM3912049 |
1422 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs759471287 | 1422 | E>Q | No |
ExAC gnomAD |
|
| rs1983570763 | 1423 | G>E | No | Ensembl | |
| rs2148531521 | 1423 | G>R | No | Ensembl | |
| rs1223964734 | 1425 | R>Q | No |
TOPMed gnomAD |
|
|
rs187930521 COSM444401 |
1425 | R>W | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed |
| TCGA novel | 1427 | K>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1428 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775539589 | 1428 | E>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1429 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs905979723 | 1429 | N>S | No |
TOPMed gnomAD |
|
| rs764039448 | 1431 | D>E | No |
ExAC gnomAD |
|
| rs977211798 | 1431 | D>Y | No | Ensembl | |
| rs761560907 | 1432 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs761560907 | 1432 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs377669374 | 1432 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs764762784 | 1434 | E>G | No |
ExAC gnomAD |
|
| rs749922516 | 1435 | W>G | No |
ExAC gnomAD |
|
| rs2148531682 | 1437 | Q>* | No | Ensembl | |
| rs1293890007 | 1439 | H>R | No | TOPMed | |
| rs549648084 | 1439 | H>Y | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs756454056 | 1442 | C>R | No |
ExAC gnomAD |
|
| rs1317970810 | 1445 | L>P | No |
TOPMed gnomAD |
|
| rs1468434862 | 1450 | V>M | No | TOPMed | |
|
rs2148556842 RCV001579321 |
1452 | N>Y | No |
ClinVar Ensembl dbSNP |
|
|
rs768735426 COSM1158230 COSM3389946 |
1460 | P>L | Variant assessed as Somatic; MODERATE impact. pancreas central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| COSM1030420 | 1460 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
RCV002462339 COSM5755653 RCV001172294 COSM5755654 rs1985063411 |
1461 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar TOPMed dbSNP |
|
rs1357531112 COSM341235 |
1461 | R>H | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1985063411 | 1461 | R>S | No | TOPMed | |
| rs1189577938 | 1465 | H>D | No | Ensembl | |
| rs747917600 | 1466 | S>R | No | ExAC | |
| rs769695219 | 1467 | G>E | No |
ExAC gnomAD |
|
| rs1242319827 | 1468 | K>Q | No | TOPMed | |
| rs772722021 | 1470 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1985067943 | 1471 | K>E | No | TOPMed | |
| TCGA novel | 1473 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1030421 | 1475 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1479 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1480 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1480 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1985069875 | 1483 | F>C | No | gnomAD | |
| rs1985070615 | 1484 | N>D | No | gnomAD | |
| rs1347687279 | 1485 | D>A | No | gnomAD | |
| COSM300378 | 1485 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs1293814714 COSM3550458 COSM3550457 |
1487 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs754410795 | 1491 | Q>H | No |
ExAC gnomAD |
|
|
rs1040261538 COSM1413918 COSM5829100 |
1493 | T>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1040261538 | 1493 | T>R | No |
TOPMed gnomAD |
|
| rs779828423 | 1495 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs780230800 | 1495 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs779828423 | 1495 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs749024844 | 1501 | T>N | No | Ensembl | |
| rs1985078128 | 1502 | D>E | No | Ensembl | |
| rs773094165 | 1502 | D>H | No |
ExAC gnomAD |
|
| rs773094165 | 1502 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1178947112 | 1503 | K>E | No | gnomAD | |
| rs748847627 | 1504 | V>A | No |
ExAC gnomAD |
|
| rs1985079996 | 1508 | K>R | No | TOPMed | |
| rs774034996 | 1512 | Q>H | No | ExAC | |
| rs1985081191 | 1513 | Y>F | No | TOPMed | |
|
COSM6161375 COSM6161376 |
1513 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3841846 COSM3841845 |
1514 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1515 | M>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1386736207 TCGA novel |
1515 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1985081582 | 1515 | M>T | No | Ensembl | |
| rs200922343 | 1519 | P>A | No |
1000Genomes ExAC gnomAD |
|
| rs1985286612 | 1519 | P>H | No | Ensembl | |
| rs200922343 | 1519 | P>T | No |
1000Genomes ExAC gnomAD |
|
| rs1269880681 | 1522 | L>V | No | gnomAD | |
| rs1602715207 | 1532 | D>A | No | Ensembl | |
| rs75569856 | 1536 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1344783696 | 1536 | D>N | No | TOPMed | |
| rs1985291299 | 1537 | E>D | No | Ensembl | |
| rs773631437 | 1537 | E>K | No |
ExAC gnomAD |
|
| rs1985293106 | 1545 | I>N | No | TOPMed | |
| rs751769880 | 1545 | I>V | No |
ExAC gnomAD |
|
|
COSM5201565 rs759839079 COSM444402 |
1547 | R>C | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM4295921 rs374602007 COSM1681947 |
1547 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs759839079 | 1547 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs756190215 | 1548 | V>I | No | ExAC | |
| rs1423613630 | 1549 | Y>C | No | gnomAD | |
| rs1985294592 | 1549 | Y>H | No | Ensembl | |
| rs1985295287 | 1550 | T>I | No | TOPMed | |
| rs777765337 | 1551 | L>I | No |
ExAC gnomAD |
|
| rs777765337 | 1551 | L>V | No |
ExAC gnomAD |
|
|
rs1408360223 COSM419248 |
1552 | R>Q | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1985296890 | 1553 | A>T | No | Ensembl | |
|
COSM6094691 COSM6094692 |
1554 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs754450226 | 1560 | T>A | No |
ExAC gnomAD |
|
| rs1011072630 | 1560 | T>I | No |
TOPMed gnomAD |
|
| rs780567832 | 1561 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs780567832 | 1561 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs202056804 | 1561 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs771546522 | 1563 | V>L | No |
ExAC gnomAD |
|
| rs771546522 | 1563 | V>M | No |
ExAC gnomAD |
|
| TCGA novel | 1565 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746235581 | 1568 | A>D | No |
ExAC gnomAD |
|
| rs1425009835 | 1570 | S>Y | No | gnomAD | |
|
COSM4847366 COSM4847367 |
1571 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772381145 | 1572 | L>H | No |
ExAC gnomAD |
|
| rs1368558188 | 1574 | I>V | No | gnomAD | |
| rs1985558977 | 1576 | T>A | No | Ensembl | |
| rs1028663752 | 1577 | E>K | No | TOPMed | |
| rs202038247 | 1578 | K>T | No |
1000Genomes ExAC gnomAD |
|
| rs1329516789 | 1581 | R>H | No |
TOPMed gnomAD |
|
| rs1445214379 | 1582 | E>K | No |
TOPMed gnomAD |
|
| rs530877301 | 1584 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1985562582 | 1584 | A>T | No | Ensembl | |
| rs530877301 | 1584 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1328630279 | 1587 | V>D | No | gnomAD | |
| rs1985564375 | 1587 | V>I | No | gnomAD | |
| rs2148566630 | 1588 | R>G | No | Ensembl | |
| rs1164637837 | 1588 | R>H | No |
TOPMed gnomAD |
|
| rs946876442 | 1589 | S>Y | No |
TOPMed gnomAD |
|
| TCGA novel | 1590 | Q>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs147520772 | 1592 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs755631618 | 1594 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs755631618 | 1594 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1259124176 | 1597 | R>W | No | gnomAD | |
| rs1391103034 | 1598 | L>V | No |
TOPMed gnomAD |
|
|
rs2148566734 COSM1030422 |
1600 | V>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs191579985 | 1601 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs758854261 | 1606 | I>L | No |
ExAC gnomAD |
|
|
rs372197768 COSM1316605 |
1607 | E>K | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ESP TOPMed gnomAD |
| rs769129278 | 1609 | K>E | No |
ExAC TOPMed gnomAD |
|
| rs1985634311 | 1609 | K>T | No | TOPMed | |
| rs1342238619 | 1610 | P>S | No | gnomAD | |
| rs1321411277 | 1611 | C>S | No | gnomAD | |
| rs781378050 | 1612 | R>Q | No |
ExAC gnomAD |
|
| rs897452417 | 1612 | R>W | No |
TOPMed gnomAD |
|
| rs1985636912 | 1613 | S>L | No | Ensembl | |
|
COSM355571 rs771953471 |
1616 | K>R | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM3550459 COSM3550460 |
1618 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs375436225 | 1619 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1985797544 | 1621 | C>Y | No | TOPMed | |
| rs1569346360 | 1627 | S>T | No | Ensembl | |
| rs1985799660 | 1628 | Q>K | No | Ensembl | |
| rs1985800506 | 1632 | T>S | No | Ensembl | |
| rs1569346384 | 1633 | K>Q | No | Ensembl | |
| rs1017841591 | 1633 | K>R | No |
TOPMed gnomAD |
|
| rs145479081 | 1634 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1318806349 | 1635 | I>M | No | TOPMed | |
| rs756507579 | 1637 | D>E | No |
ExAC gnomAD |
|
| rs753036054 | 1637 | D>G | No |
ExAC gnomAD |
|
| rs1206619692 | 1639 | L>R | No | gnomAD | |
| rs371476437 | 1642 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1751621 rs905634308 |
1645 | S>F | urinary_tract [Cosmic] | No |
cosmic curated Ensembl |
| rs79461700 | 1645 | S>P | No | Ensembl | |
| rs1985805865 | 1646 | N>S | No | Ensembl | |
| rs1985806208 | 1647 | C>G | No | Ensembl | |
| rs757221832 | 1647 | C>Y | No |
ExAC gnomAD |
|
| rs1985807306 | 1650 | F>S | No | TOPMed | |
| rs1985807633 | 1651 | I>V | No | Ensembl | |
| rs1473952903 | 1652 | R>* | No |
TOPMed gnomAD |
|
| rs1333297978 | 1652 | R>Q | No |
TOPMed gnomAD |
|
| rs1002075112 | 1655 | E>V | No |
TOPMed gnomAD |
|
| rs1429626537 | 1661 | I>M | No | gnomAD | |
| rs772113073 | 1665 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2148570448 | 1670 | S>L | No | Ensembl | |
| rs1035866881 | 1672 | D>E | No | Ensembl | |
|
COSM4101421 COSM4101420 |
1674 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1675 | L>F | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1986081355 | 1678 | T>A | No | gnomAD | |
|
COSM1681948 rs762442834 |
1678 | T>M | large_intestine [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
rs1248501456 COSM1030423 |
1681 | R>C | pancreas endometrium [Cosmic] | No |
cosmic curated 1000Genomes TOPMed gnomAD |
|
rs775912926 COSM1413919 COSM5141356 |
1681 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs569760339 | 1682 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs777159341 | 1683 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs765494185 | 1685 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1986084488 | 1685 | I>V | No | TOPMed | |
| TCGA novel | 1686 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1049059139 | 1688 | D>H | No | Ensembl | |
| TCGA novel | 1688 | D>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1689 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1325502152 | 1689 | Q>R | No | gnomAD | |
|
COSM3550463 COSM3550464 |
1690 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs201935892 | 1692 | K>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751501653 | 1693 | G>S | No |
ExAC TOPMed |
|
| rs1440879945 | 1695 | V>L | No | gnomAD | |
| rs369283779 | 1696 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 1697 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1290026665 | 1699 | L>R | No | gnomAD | |
| rs1986089324 | 1702 | H>Y | No | TOPMed | |
| rs769542325 | 1705 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1569348934 | 1705 | P>S | No | gnomAD | |
| rs1292677447 | 1706 | T>M | No | gnomAD | |
| rs1181191917 | 1707 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1393876470 | 1709 | I>T | No | gnomAD | |
| TCGA novel | 1710 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1251628654 | 1712 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs770392198 | 1712 | R>H | No |
ExAC TOPMed gnomAD |
|
|
COSM3550465 COSM3550466 |
1714 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1718 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs759096233 | 1719 | D>E | No |
ExAC gnomAD |
|
| rs372446522 | 1719 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM6094690 COSM579265 rs1986094522 COSM6094689 |
1720 | E>K | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic TOPMed |
|
rs145445183 RCV000997827 |
1721 | P>L | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
| rs762227353 | 1722 | P>Q | No |
ExAC gnomAD |
No associated diseases with Q15811
16 regional properties for Q15811
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 1579 - 1695 | IPR000008 |
| domain | Dbl homology (DH) domain | 1237 - 1423 | IPR000219 |
| domain | EH domain | 14 - 108 | IPR000261-1 |
| domain | EH domain | 214 - 310 | IPR000261-2 |
| conserved_site | Guanine-nucleotide dissociation stimulator, CDC24, conserved site | 1371 - 1396 | IPR001331 |
| domain | SH3 domain | 740 - 806 | IPR001452-1 |
| domain | SH3 domain | 913 - 971 | IPR001452-2 |
| domain | SH3 domain | 1002 - 1060 | IPR001452-3 |
| domain | SH3 domain | 1074 - 1138 | IPR001452-4 |
| domain | SH3 domain | 1155 - 1214 | IPR001452-5 |
| domain | Pleckstrin homology domain | 1440 - 1593 | IPR001849 |
| domain | EF-hand domain | 53 - 88 | IPR002048-1 |
| domain | EF-hand domain | 254 - 289 | IPR002048-2 |
| binding_site | EF-Hand 1, calcium-binding site | 66 - 78 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 267 - 279 | IPR018247-2 |
| domain | Intersectin-1, AP2 binding region | 803 - 917 | IPR032140 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR46006 | RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR AT 64C, ISOFORM A |
| PANTHER Subfamily | PTHR46006:SF9 | INTERSECTIN-2 ISOFORM X1 |
| PANTHER Protein Class |
guanyl-nucleotide exchange factor
G-protein modulator protein-binding activity modulator |
|
| PANTHER Pathway Category | No pathway information available | |
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| clathrin-coated pit | A part of the endomembrane system in the form of an invagination of a membrane upon which a clathrin coat forms, and that can be converted by vesicle budding into a clathrin-coated vesicle. Coated pits form on the plasma membrane, where they are involved in receptor-mediated selective transport of many proteins and other macromolecules across the cell membrane, in the trans-Golgi network, and on some endosomes. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| intracellular vesicle | Any vesicle that is part of the intracellular region. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| nuclear envelope | The double lipid bilayer enclosing the nucleus and separating its contents from the rest of the cytoplasm; includes the intermembrane space, a gap of width 20-40 nm (also called the perinuclear space). |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| presynaptic membrane | A specialized area of membrane of the axon terminal that faces the plasma membrane of the neuron or muscle fiber with which the axon terminal establishes a synaptic junction; many synaptic junctions exhibit structural presynaptic characteristics, such as conical, electron-dense internal protrusions, that distinguish it from the remainder of the axon plasma membrane. |
| recycling endosome | An organelle consisting of a network of tubules that functions in targeting molecules, such as receptors transporters and lipids, to the plasma membrane. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| guanyl-nucleotide exchange factor activity | Stimulates the exchange of GDP to GTP on a signaling GTPase, changing its conformation to its active form. Guanine nucleotide exchange factors (GEFs) act by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP), which is more abundant in the cell under normal cellular physiological conditions. |
| molecular adaptor activity | The binding activity of a molecule that brings together two or more molecules through a selective, non-covalent, often stoichiometric interaction, permitting those molecules to function in a coordinated way. |
| proline-rich region binding | Binding to a proline-rich region, i.e. a region that contains a high proportion of proline residues, in a protein. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| clathrin-dependent synaptic vesicle endocytosis | Clathrin-dependent endocytosis of presynaptic membrane regions comprising synaptic vesicles' membrane constituents. This is a relatively slow process occurring in the range of tens of seconds. |
| endosomal transport | The directed movement of substances mediated by an endosome, a membrane-bounded organelle that carries materials enclosed in the lumen or located in the endosomal membrane. |
| exocytosis | A process of secretion by a cell that results in the release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle. Exocytosis can occur either by full fusion, when the vesicle collapses into the plasma membrane, or by a kiss-and-run mechanism that involves the formation of a transient contact, a pore, between a granule (for exemple of chromaffin cells) and the plasma membrane. The latter process most of the time leads to only partial secretion of the granule content. Exocytosis begins with steps that prepare vesicles for fusion with the membrane (tethering and docking) and ends when molecules are secreted from the cell. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| protein localization | Any process in which a protein is transported to, or maintained in, a specific location. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| regulation of small GTPase mediated signal transduction | Any process that modulates the frequency, rate or extent of small GTPase mediated signal transduction. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| E1BKY4 | ITSN1 | Intersectin 1 | Bos taurus (Bovine) | SS |
| A0A2I2U1K8 | ITSN1 | Intersectin 1 | Felis catus (Cat) (Felis silvestris catus) | SS |
| A0A2I3T257 | ITSN1 | Intersectin 1 | Pan troglodytes (Chimpanzee) | SS |
| Q9NZM3 | ITSN2 | Intersectin-2 | Homo sapiens (Human) | SS |
| Q8NFH8 | REPS2 | RalBP1-associated Eps domain-containing protein 2 | Homo sapiens (Human) | PR |
| Q9Z0R6 | Itsn2 | Intersectin-2 | Mus musculus (Mouse) | SS |
| Q9Z0R4 | Itsn1 | Intersectin-1 | Mus musculus (Mouse) | EV |
| Q9WVE9 | Itsn1 | Intersectin-1 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAQFPTPFGG | SLDIWAITVE | ERAKHDQQFH | SLKPISGFIT | GDQARNFFFQ | SGLPQPVLAQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IWALADMNND | GRMDQVEFSI | AMKLIKLKLQ | GYQLPSALPP | VMKQQPVAIS | SAPAFGMGGI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ASMPPLTAVA | PVPMGSIPVV | GMSPTLVSSV | PTAAVPPLAN | GAPPVIQPLP | AFAHPAATLP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| KSSSFSRSGP | GSQLNTKLQK | AQSFDVASVP | PVAEWAVPQS | SRLKYRQLFN | SHDKTMSGHL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TGPQARTILM | QSSLPQAQLA | SIWNLSDIDQ | DGKLTAEEFI | LAMHLIDVAM | SGQPLPPVLP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PEYIPPSFRR | VRSGSGISVI | SSTSVDQRLP | EEPVLEDEQQ | QLEKKLPVTF | EDKKRENFER |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GNLELEKRRQ | ALLEQQRKEQ | ERLAQLERAE | QERKERERQE | QERKRQLELE | KQLEKQRELE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RQREEERRKE | IERREAAKRE | LERQRQLEWE | RNRRQELLNQ | RNKEQEDIVV | LKAKKKTLEF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ELEALNDKKH | QLEGKLQDIR | CRLTTQRQEI | ESTNKSRELR | IAEITHLQQQ | LQESQQMLGR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LIPEKQILND | QLKQVQQNSL | HRDSLVTLKR | ALEAKELARQ | HLRDQLDEVE | KETRSKLQEI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| DIFNNQLKEL | REIHNKQQLQ | KQKSMEAERL | KQKEQERKII | ELEKQKEEAQ | RRAQERDKQW |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LEHVQQEDEH | QRPRKLHEEE | KLKREESVKK | KDGEEKGKQE | AQDKLGRLFH | QHQEPAKPAV |
| 730 | 740 | 750 | 760 | 770 | 780 |
| QAPWSTAEKG | PLTISAQENV | KVVYYRALYP | FESRSHDEIT | IQPGDIVMVK | GEWVDESQTG |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EPGWLGGELK | GKTGWFPANY | AEKIPENEVP | APVKPVTDST | SAPAPKLALR | ETPAPLAVTS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SEPSTTPNNW | ADFSSTWPTS | TNEKPETDNW | DAWAAQPSLT | VPSAGQLRQR | SAFTPATATG |
| 910 | 920 | 930 | 940 | 950 | 960 |
| SSPSPVLGQG | EKVEGLQAQA | LYPWRAKKDN | HLNFNKNDVI | TVLEQQDMWW | FGEVQGQKGW |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| FPKSYVKLIS | GPIRKSTSMD | SGSSESPASL | KRVASPAAKP | VVSGEEFIAM | YTYESSEQGD |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| LTFQQGDVIL | VTKKDGDWWT | GTVGDKAGVF | PSNYVRLKDS | EGSGTAGKTG | SLGKKPEIAQ |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| VIASYTATGP | EQLTLAPGQL | ILIRKKNPGG | WWEGELQARG | KKRQIGWFPA | NYVKLLSPGT |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| SKITPTEPPK | STALAAVCQV | IGMYDYTAQN | DDELAFNKGQ | IINVLNKEDP | DWWKGEVNGQ |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| VGLFPSNYVK | LTTDMDPSQQ | WCSDLHLLDM | LTPTERKRQG | YIHELIVTEE | NYVNDLQLVT |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| EIFQKPLMES | ELLTEKEVAM | IFVNWKELIM | CNIKLLKALR | VRKKMSGEKM | PVKMIGDILS |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| AQLPHMQPYI | RFCSRQLNGA | ALIQQKTDEA | PDFKEFVKRL | AMDPRCKGMP | LSSFILKPMQ |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| RVTRYPLIIK | NILENTPENH | PDHSHLKHAL | EKAEELCSQV | NEGVREKENS | DRLEWIQAHV |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| QCEGLSEQLV | FNSVTNCLGP | RKFLHSGKLY | KAKSNKELYG | FLFNDFLLLT | QITKPLGSSG |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| TDKVFSPKSN | LQYKMYKTPI | FLNEVLVKLP | TDPSGDEPIF | HISHIDRVYT | LRAESINERT |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| AWVQKIKAAS | ELYIETEKKK | REKAYLVRSQ | RATGIGRLMV | NVVEGIELKP | CRSHGKSNPY |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| CEVTMGSQCH | ITKTIQDTLN | PKWNSNCQFF | IRDLEQEVLC | ITVFERDQFS | PDDFLGRTEI |
| 1690 | 1700 | 1710 | 1720 | ||
| RVADIKKDQG | SKGPVTKCLL | LHEVPTGEIV | VRLDLQLFDE | P |