Q15375
Gene name |
EPHA7 (EHK3, HEK11) |
Protein name |
Ephrin type-A receptor 7 |
Names |
EC 2.7.10.1 , EPH homology kinase 3 , EHK-3 , EPH-like kinase 11 , EK11 , hEK11 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2045 |
EC number |
2.7.10.1: Protein-tyrosine kinases |
Protein Class |
|
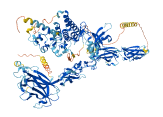
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
633-894 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
775-801 (Activation loop from InterPro)
Target domain |
633-894 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
Autoinhibited structure

Activated structure

8 structures for Q15375
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2REI | X-ray | 160 A | A | 590-899 | PDB |
| 3DKO | X-ray | 200 A | A | 590-899 | PDB |
| 3H8M | X-ray | 210 A | A/B | 919-990 | PDB |
| 3NRU | X-ray | 230 A | A/B/C/D/E/F/G/H/I/J/K/L | 32-204 | PDB |
| 7EEC | X-ray | 310 A | A | 599-899 | PDB |
| 7EED | X-ray | 305 A | A | 599-899 | PDB |
| 7EEF | X-ray | 260 A | A | 599-899 | PDB |
| AF-Q15375-F1 | Predicted | AlphaFoldDB |
1291 variants for Q15375
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1273287575 | 3 | F>L | No |
TOPMed gnomAD |
|
| rs202112730 | 4 | Q>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| TCGA novel | 4 | Q>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1337239729 | 6 | R>L | No |
TOPMed gnomAD |
|
| rs1337239729 | 6 | R>P | No |
TOPMed gnomAD |
|
| rs746638787 | 8 | P>S | No |
TOPMed gnomAD |
|
| rs1779414064 | 9 | S>* | No | TOPMed | |
|
COSM3630632 COSM4894664 |
10 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772524127 | 10 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA |
| rs1199857559 | 12 | I>L | No | gnomAD | |
| rs140401119 | 12 | I>T | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 13 | L>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1779413323 | 13 | L>F | No | Ensembl | |
| rs779195088 | 13 | L>S | No |
ExAC gnomAD |
|
| rs146742012 | 15 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1779412834 | 16 | I>T | No | TOPMed | |
| rs1258617697 | 17 | W>S | No | gnomAD | |
| rs1216196756 | 18 | L>P | No |
TOPMed gnomAD |
|
| rs1779412241 | 19 | L>H | No | TOPMed | |
| rs149722351 | 20 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1198612981 | 20 | R>H | No |
TOPMed gnomAD |
|
| rs1373981234 | 23 | H>R | No |
TOPMed gnomAD |
|
| rs752294486 | 24 | T>A | No |
ExAC gnomAD |
|
| rs200143358 | 24 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs752294486 | 24 | T>P | No |
ExAC gnomAD |
|
| rs200143358 | 24 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs529519500 | 25 | G>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs529519500 | 25 | G>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs370009055 | 25 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1401739411 | 26 | E>Q | No | gnomAD | |
| rs773296055 | 27 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1779409784 | 29 | A>T | No | gnomAD | |
|
COSM5107681 COSM269112 |
30 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1389987827 | 31 | K>R | No |
TOPMed gnomAD |
|
| rs1389987827 | 31 | K>T | No |
TOPMed gnomAD |
|
| rs775107464 | 33 | V>I | No | Ensembl | |
| rs1192639917 | 35 | L>R | No | gnomAD | |
| rs750219466 | 38 | S>F | No |
ExAC gnomAD |
|
| rs1289923558 | 41 | Q>H | No | gnomAD | |
|
COSM4992803 COSM4992802 rs1339057136 |
44 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs140813099 | 46 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs140813099 | 46 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4899208 COSM3630631 |
48 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1292019841 | 49 | S>C | No | gnomAD | |
|
COSM1697651 COSM4897316 |
49 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 49 | S>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1779140955 | 50 | S>F | No | TOPMed | |
| rs1232604941 | 51 | P>S | No |
TOPMed gnomAD |
|
| rs1268640001 | 52 | P>S | No |
TOPMed gnomAD |
|
| rs1779140428 | 53 | N>S | No | Ensembl | |
| rs867992353 | 57 | E>K | No | Ensembl | |
| rs766551054 | 58 | I>V | No |
ExAC gnomAD |
|
| rs763098772 | 61 | L>F | No |
ExAC gnomAD |
|
| rs1778957821 | 62 | D>G | No | TOPMed | |
| TCGA novel | 63 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374771258 | 65 | Y>C | No |
ESP ExAC gnomAD |
|
| rs750661621 | 65 | Y>H | No |
ExAC gnomAD |
|
| rs1238158906 | 66 | T>S | No | gnomAD | |
|
COSM1082677 rs889062730 COSM4872472 |
67 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs889062730 | 67 | P>Q | No | gnomAD | |
|
COSM4407906 rs1300929218 COSM126676 |
69 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1300929218 | 69 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
rs1465346253 COSM3922264 COSM35826 |
69 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| TCGA novel | 70 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1778955869 | 71 | Y>* | No | TOPMed | |
| rs1778955724 | 72 | Q>E | No | TOPMed | |
| TCGA novel | 73 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761300271 | 77 | M>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 80 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200883289 | 80 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM3630630 COSM4898953 |
81 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM5575918 COSM5575919 rs745460482 |
82 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs774165896 | 83 | N>S | No |
ExAC gnomAD |
|
| rs570778929 | 86 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs770806616 COSM4782877 COSM1446507 |
86 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1036535390 | 87 | T>S | No | TOPMed | |
| rs756003702 | 88 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs1214746243 | 88 | N>T | No |
TOPMed gnomAD |
|
|
COSM3630629 COSM4539403 |
89 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748124897 | 90 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1778952552 | 91 | S>F | No | Ensembl | |
| rs1241267544 | 93 | G>D | No | gnomAD | |
| rs779944312 | 94 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs981923012 | 94 | N>S | No |
TOPMed gnomAD |
|
|
COSM1446506 COSM4786528 |
95 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127994969 | 96 | Q>L | No | Ensembl | |
| rs1778951148 | 98 | I>T | No | Ensembl | |
| rs1314436572 | 99 | F>S | No | gnomAD | |
| rs1379307134 | 100 | V>L | No | gnomAD | |
| rs1778950274 | 103 | K>Q | No | TOPMed | |
| rs1158714216 | 104 | F>C | No | gnomAD | |
| rs754137746 | 106 | L>V | No |
ExAC gnomAD |
|
|
rs1778949138 COSM70629 COSM5608520 |
108 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1778949003 | 114 | G>R | No | TOPMed | |
|
COSM4357534 COSM3178904 |
117 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764585634 | 118 | T>A | No |
ExAC TOPMed gnomAD |
|
|
COSM743811 COSM4861114 |
121 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776073346 | 124 | N>H | No |
ExAC gnomAD |
|
| rs767086353 | 125 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs759146981 | 126 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1469665299 | 126 | Y>H | No | gnomAD | |
| rs774043406 | 127 | Y>S | No |
ExAC gnomAD |
|
| TCGA novel | 131 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1232627325 | 132 | Y>F | No | gnomAD | |
| rs1355988571 | 133 | D>E | No | gnomAD | |
| rs1778947291 | 133 | D>N | No | Ensembl | |
| rs1778947015 | 134 | T>S | No | gnomAD | |
| rs2127994905 | 136 | R>G | No | Ensembl | |
| rs1251015381 | 136 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1397911143 | 137 | N>D | No | gnomAD | |
|
rs1318584442 COSM1082675 COSM4864989 |
138 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
|
VAR_022105 rs2278107 |
138 | I>V | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM4864872 COSM1082673 |
140 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3749990 COSM4782533 |
140 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs548216830 | 141 | N>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2127994890 | 141 | N>T | No | Ensembl | |
| rs200025945 | 142 | L>I | No |
1000Genomes ExAC gnomAD |
|
| rs1778945237 | 142 | L>R | No | TOPMed | |
| rs1240239966 | 143 | Y>C | No | TOPMed | |
| rs781143231 | 143 | Y>H | No |
ExAC gnomAD |
|
| rs2127994879 | 144 | V>L | No | Ensembl | |
| TCGA novel | 149 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM1446502 COSM4785916 |
151 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4906240 COSM3630627 |
153 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1484097577 | 153 | E>Q | No | TOPMed | |
| rs779024743 | 157 | Q>P | No |
ExAC gnomAD |
|
|
COSM4922579 COSM4922578 rs1286516366 |
158 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1778942474 | 159 | D>Y | No | Ensembl | |
| rs1582691994 | 165 | M>V | No | Ensembl | |
|
COSM3941882 rs754092318 |
166 | K>R | oesophagus [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1778941480 | 169 | T>I | No | Ensembl | |
| VAR_042150 | 170 | E>K | a colorectal adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
| rs1241523900 | 172 | R>G | No |
TOPMed gnomAD |
|
| rs1444159555 | 173 | E>G | No | TOPMed | |
| rs1562167396 | 173 | E>K | No | TOPMed | |
| rs753104602 | 175 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1778939989 | 176 | P>T | No | Ensembl | |
| rs770996678 | 177 | L>S | No | Ensembl | |
| rs1778939683 | 178 | S>F | No | TOPMed | |
| rs1323379573 | 179 | K>E | No | Ensembl | |
| TCGA novel | 180 | K>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3630626 COSM4891761 |
181 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1778938900 | 183 | Y>C | No | TOPMed | |
| rs1418432799 | 183 | Y>H | No |
TOPMed gnomAD |
|
|
COSM4861184 rs760168204 COSM743812 |
184 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs760168204 | 184 | L>V | No |
ExAC gnomAD |
|
| rs765994473 | 187 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs2127994778 | 189 | V>A | No | Ensembl | |
| rs2127994778 | 189 | V>E | No | Ensembl | |
| rs1395960850 | 189 | V>I | No |
TOPMed gnomAD |
|
| rs2127994773 | 190 | G>V | No | Ensembl | |
| TCGA novel | 190 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 192 | C>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127994760 | 192 | C>W | No | Ensembl | |
| rs2127994763 | 192 | C>Y | No | Ensembl | |
| rs1467878949 | 193 | I>L | No | TOPMed | |
| rs1467878949 | 193 | I>V | No | TOPMed | |
|
COSM743813 COSM4861374 rs2127994750 |
194 | A>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127994750 | 194 | A>S | No | Ensembl | |
| rs1329911899 | 195 | L>F | No | gnomAD | |
| rs944145438 | 196 | V>F | No | Ensembl | |
| rs944145438 | 196 | V>I | No | Ensembl | |
| rs944145438 | 196 | V>L | No | Ensembl | |
| rs2127994735 | 197 | S>F | No | Ensembl | |
| rs2127994725 | 198 | V>A | No | Ensembl | |
| rs772940573 | 198 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs2127994717 | 199 | K>I | No | Ensembl | |
| TCGA novel | 199 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1169985049 | 200 | V>A | No | gnomAD | |
| rs1169985049 | 200 | V>E | No | gnomAD | |
| rs2127994703 | 201 | Y>* | No | Ensembl | |
| rs1778936680 | 201 | Y>C | No | TOPMed | |
| rs1778936680 | 201 | Y>F | No | TOPMed | |
| rs2127994692 | 202 | Y>* | No | Ensembl | |
| rs1446975884 | 202 | Y>F | No |
TOPMed gnomAD |
|
| rs2127994700 | 202 | Y>N | No | Ensembl | |
|
COSM3831252 COSM3831251 |
203 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs377435864 | 203 | K>M | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1778936323 COSM256864 |
203 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
TOPMed NCI-TCGA Cosmic |
| rs377435864 | 203 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs2127994673 | 204 | K>N | No | Ensembl | |
| rs2127994678 | 204 | K>R | No | Ensembl | |
| rs2127994668 | 205 | C>F | No | Ensembl | |
| rs1373965971 | 205 | C>W | No | TOPMed | |
| rs2127994657 | 206 | W>C | No | Ensembl | |
|
rs2127994662 COSM70628 |
206 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127994663 | 206 | W>R | No | Ensembl | |
| rs1778936009 | 207 | S>C | No | TOPMed | |
|
COSM35538 rs1778936009 COSM4905990 |
207 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs2127994647 | 208 | I>F | No | Ensembl | |
| rs2127994644 | 208 | I>N | No | Ensembl | |
| rs776462833 | 209 | I>T | No |
ExAC gnomAD |
|
| rs761641519 | 209 | I>V | No |
ExAC gnomAD |
|
|
COSM3831250 COSM3831249 |
210 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127994625 | 210 | E>D | No | Ensembl | |
| rs2127994628 | 210 | E>G | No | Ensembl | |
| rs2127994628 | 210 | E>V | No | Ensembl | |
| rs2127994617 | 211 | N>I | No | Ensembl | |
| rs2127994612 | 211 | N>K | No | Ensembl | |
| rs2127994617 | 211 | N>S | No | Ensembl | |
| rs2127994624 | 211 | N>Y | No | Ensembl | |
| rs2127994607 | 212 | L>F | No | Ensembl | |
| rs2127994600 | 213 | A>G | No | Ensembl | |
| rs2127994604 | 213 | A>P | No | Ensembl | |
| rs1778934817 | 214 | I>M | No | Ensembl | |
| rs958285932 | 214 | I>N | No | TOPMed | |
| rs532369845 | 214 | I>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1778934644 | 216 | P>A | No | Ensembl | |
| rs1778934644 | 216 | P>S | No | Ensembl | |
| rs2127994583 | 217 | D>E | No | Ensembl | |
| rs2127994589 | 217 | D>H | No | Ensembl | |
| rs2127994576 | 218 | T>I | No | Ensembl | |
| rs2127994576 | 218 | T>R | No | Ensembl | |
| rs2127994580 | 218 | T>S | No | Ensembl | |
| rs2127994570 | 220 | T>S | No | Ensembl | |
| rs2127994566 | 220 | T>S | No | Ensembl | |
| rs1207755122 | 221 | G>A | No | TOPMed | |
| rs2127994558 | 221 | G>C | No | Ensembl | |
| rs1207755122 | 221 | G>D | No | TOPMed | |
| rs2127994558 | 221 | G>R | No | Ensembl | |
| rs1207755122 | 221 | G>V | No | TOPMed | |
| rs1778934322 | 222 | S>* | No | TOPMed | |
| rs1778934322 | 222 | S>L | No | TOPMed | |
| rs2127994550 | 222 | S>T | No | Ensembl | |
| rs778894011 | 223 | E>D | No |
ExAC gnomAD |
|
| rs2127994547 | 223 | E>K | No | Ensembl | |
| rs2127994547 | 223 | E>Q | No | Ensembl | |
| rs2127994545 | 223 | E>V | No | Ensembl | |
| TCGA novel | 225 | S>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM3630625 COSM4891873 |
225 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127994525 | 226 | S>C | No | Ensembl | |
| rs2127994525 | 226 | S>F | No | Ensembl | |
| rs2127994529 | 226 | S>P | No | Ensembl | |
| rs2127994529 | 226 | S>T | No | Ensembl | |
| rs923934722 | 227 | L>V | No |
TOPMed gnomAD |
|
| rs2127994518 | 228 | V>F | No | Ensembl | |
| rs2127994518 | 228 | V>L | No | Ensembl | |
| rs1216099858 | 229 | E>* | No | gnomAD | |
| rs1342512195 | 229 | E>D | No | gnomAD | |
| rs1778932726 | 229 | E>G | No | Ensembl | |
| rs1216099858 | 229 | E>K | No | gnomAD | |
| rs1216099858 | 229 | E>Q | No | gnomAD | |
| rs1778932726 | 229 | E>V | No | Ensembl | |
| rs2127994500 | 230 | V>D | No | Ensembl | |
| rs2127994500 | 230 | V>G | No | Ensembl | |
| rs1050292701 | 230 | V>I | No |
TOPMed gnomAD |
|
| rs1050292701 | 230 | V>L | No |
TOPMed gnomAD |
|
|
COSM4357512 rs749418654 COSM1718750 |
231 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs749418654 | 231 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs778087526 | 231 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs778087526 | 231 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2127994467 | 232 | G>A | No | Ensembl | |
| rs2127994467 | 232 | G>E | No | Ensembl | |
|
rs2127994473 VAR_042151 |
232 | G>R | a metastatic melanoma sample; somatic mutation [UniProt] | No |
Ensembl UniProt |
| rs2127994467 | 232 | G>V | No | Ensembl | |
| rs753087941 | 233 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs753087941 | 233 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs2127994462 | 233 | T>S | No | Ensembl | |
| rs1345265318 | 234 | C>* | No | gnomAD | |
| rs2127994455 | 234 | C>R | No | Ensembl | |
| rs2127994453 | 234 | C>S | No | Ensembl | |
| rs1345265318 | 234 | C>W | No | gnomAD | |
| rs2127994445 | 235 | V>D | No | Ensembl | |
| rs2127994445 | 235 | V>G | No | Ensembl | |
| rs1319640969 | 235 | V>I | No | gnomAD | |
| rs1319640969 | 235 | V>L | No | gnomAD | |
| rs2127994437 | 236 | S>C | No | Ensembl | |
| rs2127994437 | 236 | S>G | No | Ensembl | |
| rs2127994434 | 236 | S>N | No | Ensembl | |
| rs2127994429 | 236 | S>R | No | Ensembl | |
| rs2127994434 | 236 | S>T | No | Ensembl | |
| rs2127994426 | 237 | S>R | No | Ensembl | |
| rs2127994419 | 238 | A>E | No | Ensembl | |
| rs2127994419 | 238 | A>G | No | Ensembl | |
| rs2127994424 | 238 | A>T | No | Ensembl | |
| rs2127994419 | 238 | A>V | No | Ensembl | |
| rs2127994410 | 239 | E>K | No | Ensembl | |
| rs2127994408 | 240 | E>K | No | Ensembl | |
| rs2127994408 | 240 | E>Q | No | Ensembl | |
| rs1778930410 | 241 | E>A | No |
TOPMed gnomAD |
|
| rs374346880 | 241 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1465849290 | 241 | E>K | No |
TOPMed gnomAD |
|
| rs1465849290 | 241 | E>Q | No |
TOPMed gnomAD |
|
| rs370195460 | 242 | A>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs370195460 | 242 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs2127994393 | 242 | A>P | No | Ensembl | |
| rs370195460 | 242 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs2127994379 | 243 | E>G | No | Ensembl | |
| rs2127994383 | 243 | E>K | No | Ensembl | |
| rs2127994383 | 243 | E>Q | No | Ensembl | |
| rs2127994375 | 244 | N>D | No | Ensembl | |
| rs2127994375 | 244 | N>H | No | Ensembl | |
| rs147242531 | 244 | N>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs989431209 | 244 | N>S | No | gnomAD | |
| rs2127994375 | 244 | N>Y | No | Ensembl | |
| rs2127994365 | 245 | A>D | No | Ensembl | |
| rs2127994365 | 245 | A>G | No | Ensembl | |
| rs41273629 | 245 | A>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs41273629 | 245 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs2127994365 TCGA novel |
245 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1455705870 | 246 | P>A | No |
TOPMed gnomAD |
|
| rs2127994356 | 246 | P>H | No | Ensembl | |
| rs2127994356 | 246 | P>R | No | Ensembl | |
| rs1455705870 | 246 | P>T | No |
TOPMed gnomAD |
|
| rs953155139 | 247 | R>G | No |
TOPMed gnomAD |
|
| rs2127994345 | 247 | R>K | No | Ensembl | |
| rs2127994345 | 247 | R>M | No | Ensembl | |
| rs2127994342 | 247 | R>S | No | Ensembl | |
| rs761433457 | 248 | M>I | No |
ExAC gnomAD |
|
| rs2127994334 | 248 | M>K | No | Ensembl | |
| rs2127994334 | 248 | M>R | No | Ensembl | |
| rs2127994339 | 248 | M>V | No | Ensembl | |
| rs2127994330 | 249 | H>N | No | Ensembl | |
| rs1219521062 | 249 | H>P | No | TOPMed | |
| rs1469620212 | 249 | H>Q | No | gnomAD | |
| rs1219521062 | 249 | H>R | No | TOPMed | |
| rs2127994330 | 249 | H>Y | No | Ensembl | |
| rs1233372560 | 250 | C>* | No | gnomAD | |
| rs2127994324 | 250 | C>F | No | Ensembl | |
| rs2127994328 | 250 | C>S | No | Ensembl | |
| rs2127994324 | 250 | C>S | No | Ensembl | |
| rs1233372560 | 250 | C>W | No | gnomAD | |
| rs2127994324 | 250 | C>Y | No | Ensembl | |
| rs1206466692 | 251 | S>N | No |
TOPMed gnomAD |
|
| rs2127994318 | 251 | S>R | No | Ensembl | |
| rs1206466692 | 251 | S>T | No |
TOPMed gnomAD |
|
| rs2127994308 | 252 | A>E | No | Ensembl | |
| rs2127994308 | 252 | A>G | No | Ensembl | |
| rs2127994316 | 252 | A>S | No | Ensembl | |
| rs2127994316 | 252 | A>T | No | Ensembl | |
| rs2127994308 | 252 | A>V | No | Ensembl | |
| rs2127994303 | 253 | E>D | No | Ensembl | |
| rs2127994299 | 255 | E>* | No | Ensembl | |
| rs2127994297 | 255 | E>G | No | Ensembl | |
|
COSM743814 COSM4862828 rs2127994299 |
255 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1778927708 | 256 | W>* | No | Ensembl | |
| rs2127994290 | 256 | W>C | No | Ensembl | |
| rs2127994296 | 256 | W>R | No | Ensembl | |
| rs2127994285 | 257 | L>* | No | Ensembl | |
| rs2127994287 | 257 | L>I | No | Ensembl | |
| rs1436208211 | 258 | V>E | No | Ensembl | |
| rs2127994282 | 258 | V>L | No | Ensembl | |
|
rs2127994273 TCGA novel |
259 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs2127994269 | 259 | P>H | No | Ensembl | |
| rs2127994269 | 259 | P>L | No | Ensembl | |
| rs2127994269 | 259 | P>R | No | Ensembl | |
|
rs2127994273 TCGA novel |
259 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs2127994273 | 259 | P>T | No | Ensembl | |
| rs1306506164 | 260 | I>T | No | gnomAD | |
| rs1349574955 | 260 | I>V | No |
TOPMed gnomAD |
|
| rs2127994251 | 261 | G>R | No | Ensembl | |
| rs140191777 | 264 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1374774567 | 267 | A>E | No | gnomAD | |
|
COSM4903999 COSM3630622 |
270 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749356915 | 271 | Q>P | No | ExAC | |
| rs770043619 | 273 | G>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1429079813 | 273 | G>A | No |
TOPMed gnomAD |
|
|
COSM4747635 COSM4747634 |
273 | G>E | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770043619 | 273 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1429079813 | 273 | G>V | No |
TOPMed gnomAD |
|
| rs748445630 | 274 | D>E | No |
ExAC gnomAD |
|
| rs1313843472 | 274 | D>G | No |
TOPMed gnomAD |
|
| rs990509658 | 275 | T>A | No | gnomAD | |
| rs1778924177 | 277 | E>K | No | Ensembl | |
| rs1215429530 | 278 | P>H | No |
TOPMed gnomAD |
|
|
rs2278106 VAR_022106 RCV001636173 |
278 | P>S | No |
ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM4813795 COSM451945 |
279 | C>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1364312999 | 279 | C>Y | No | gnomAD | |
| rs778426683 | 280 | G>D | No |
ExAC gnomAD |
|
| rs149408123 | 281 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1056840976 | 281 | R>H | No |
TOPMed gnomAD |
|
| rs1056840976 | 281 | R>L | No |
TOPMed gnomAD |
|
| rs1056840976 | 281 | R>P | No |
TOPMed gnomAD |
|
| rs1562122391 | 282 | G>E | No | Ensembl | |
| TCGA novel | 282 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127947049 | 283 | F>L | No | Ensembl | |
| rs755813766 | 284 | Y>C | No |
ExAC gnomAD |
|
| rs202168283 | 284 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1169442219 | 285 | K>E | No | gnomAD | |
| rs2127947028 | 287 | S>F | No | Ensembl | |
| rs767281324 | 288 | S>F | No |
ExAC gnomAD |
|
|
COSM4812231 COSM421199 |
289 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127947020 | 289 | Q>E | No | Ensembl | |
| rs1191295608 | 289 | Q>R | No | gnomAD | |
| rs1562122316 | 290 | D>G | No | Ensembl | |
| rs1313085656 | 290 | D>N | No | gnomAD | |
|
COSM3876210 COSM3876209 |
290 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1467643679 | 291 | L>H | No | gnomAD | |
|
COSM3922259 COSM3922258 rs760111731 |
292 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs760111731 | 292 | Q>E | No | Ensembl | |
| rs1160114110 | 292 | Q>R | No | TOPMed | |
| rs1776062537 | 294 | S>F | No | gnomAD | |
| rs375147203 | 295 | R>C | No |
ESP ExAC gnomAD |
|
| rs765322526 | 295 | R>H | No |
ExAC gnomAD |
|
| TCGA novel | 295 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs938343635 | 297 | P>L | No | TOPMed | |
| TCGA novel | 297 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776903613 | 298 | T>A | No |
ExAC gnomAD |
|
| rs776903613 | 298 | T>P | No |
ExAC gnomAD |
|
| rs1205658610 | 299 | H>Y | No |
TOPMed gnomAD |
|
| rs1309546660 | 302 | S>P | No | gnomAD | |
| rs1776061153 | 303 | D>N | No | Ensembl | |
|
COSM4832139 COSM4832138 |
305 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768989326 | 306 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs41273627 | 306 | G>S | No | Ensembl | |
| rs768989326 | 306 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs909460037 | 309 | R>G | No | Ensembl | |
| rs1233763479 | 310 | C>F | No | gnomAD | |
| TCGA novel | 311 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747304855 | 311 | E>K | No |
ExAC gnomAD |
|
|
COSM6107501 COSM6107500 |
312 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs113701581 | 314 | D>E | No | ExAC | |
| rs1305662700 | 316 | Y>H | No |
TOPMed gnomAD |
|
| rs1305662700 | 316 | Y>N | No |
TOPMed gnomAD |
|
|
TCGA novel rs1776059879 |
318 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs772334549 | 318 | R>T | No |
ExAC TOPMed gnomAD |
|
| rs138876328 | 319 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs779315809 | 319 | A>V | No |
ExAC gnomAD |
|
| rs757798599 | 320 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs748773525 | 321 | S>Y | No |
ExAC gnomAD |
|
| rs1582588608 | 322 | D>A | No | Ensembl | |
| rs1171397060 | 322 | D>E | No |
TOPMed gnomAD |
|
| rs1453370957 | 323 | P>L | No | gnomAD | |
| rs1395209134 | 324 | P>L | No | gnomAD | |
|
COSM4786342 COSM1446498 |
324 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3630620 COSM4891821 |
324 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1776057623 | 325 | Y>C | No | gnomAD | |
| rs1194925755 | 325 | Y>H | No | gnomAD | |
| rs1776057084 | 326 | V>D | No | gnomAD | |
| rs150543233 | 326 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1776056576 | 329 | T>R | No |
TOPMed gnomAD |
|
| rs1413927391 | 330 | R>K | No |
TOPMed gnomAD |
|
| TCGA novel | 331 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200873598 | 331 | P>T | No | Ensembl | |
| TCGA novel | 332 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs892457234 | 335 | P>S | No | Ensembl | |
| rs1380130381 | 336 | Q>E | No | gnomAD | |
| rs1158620225 | 339 | I>F | No |
TOPMed gnomAD |
|
| rs1158620225 | 339 | I>V | No |
TOPMed gnomAD |
|
| rs1775981244 | 341 | N>D | No | gnomAD | |
| rs758347037 | 341 | N>I | No |
ExAC gnomAD |
|
| rs750403324 | 343 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs1164758223 | 344 | Q>R | No | gnomAD | |
| rs1475740786 | 346 | T>A | No | gnomAD | |
| rs764217573 | 347 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1775980305 | 348 | S>G | No | Ensembl | |
|
COSM4899277 COSM3630619 |
350 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1191331342 | 353 | P>S | No | gnomAD | |
| rs1191331342 | 353 | P>T | No | gnomAD | |
| rs2127944970 | 355 | A>E | No | Ensembl | |
| rs2127944970 | 355 | A>V | No | Ensembl | |
| rs1775979799 | 357 | N>D | No | TOPMed | |
| rs1582585734 | 357 | N>S | No | Ensembl | |
| rs759223701 | 359 | G>E | No | Ensembl | |
| rs145247136 | 361 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774694250 | 363 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1428092925 | 364 | T>S | No | Ensembl | |
| rs201390062 | 364 | T>S | No |
ESP TOPMed gnomAD |
|
| rs1302457908 | 366 | R>K | No | TOPMed | |
| rs763391728 | 366 | R>S | No |
ExAC gnomAD |
|
| rs140214296 | 368 | L>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 368 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 370 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs138769019 | 371 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs138769019 | 371 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
VAR_036090 rs371089003 |
371 | R>W | a colorectal cancer sample; somatic mutation [UniProt] | No |
UniProt ESP ExAC TOPMed dbSNP gnomAD |
| rs780813699 | 372 | C>Y | No |
ExAC gnomAD |
|
| rs1391562093 | 373 | S>C | No |
TOPMed gnomAD |
|
| rs772199348 | 373 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs1391562093 | 373 | S>R | No |
TOPMed gnomAD |
|
| rs772199348 | 373 | S>T | No |
ExAC TOPMed gnomAD |
|
|
COSM4838051 COSM4838050 |
374 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1228469813 | 375 | E>G | No |
TOPMed gnomAD |
|
| rs779939317 | 376 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs779939317 | 376 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs758293765 | 377 | G>D | No |
ExAC gnomAD |
|
| rs778960808 | 378 | E>K | No | ExAC | |
| rs778960808 | 378 | E>Q | No | ExAC | |
|
COSM6107502 COSM6107503 |
379 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs930359353 | 380 | V>I | No | Ensembl | |
| rs1475247277 | 383 | G>R | No |
TOPMed gnomAD |
|
| rs1775974480 | 384 | S>N | No | TOPMed | |
| rs757264200 | 385 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs757264200 | 385 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs918912377 | 385 | N>K | No |
TOPMed gnomAD |
|
| rs1252412601 | 386 | I>T | No | gnomAD | |
| rs1452664406 | 386 | I>V | No | gnomAD | |
| rs1355103404 | 387 | G>R | No | TOPMed | |
| rs1240393280 | 389 | M>I | No | TOPMed | |
| rs1218588481 | 389 | M>K | No |
TOPMed gnomAD |
|
| rs752702243 | 389 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1775972976 | 391 | Q>K | No | Ensembl | |
| rs767702045 | 392 | Q>E | No |
ExAC gnomAD |
|
|
COSM743815 COSM4859467 |
392 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4905765 COSM3630616 |
394 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs971828200 | 395 | L>V | No |
TOPMed gnomAD |
|
| COSM272642 | 396 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1775972514 | 396 | E>A | No | TOPMed | |
| rs1182610695 | 397 | D>E | No | TOPMed | |
| rs755089288 | 397 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1168434262 | 399 | Y>C | No | gnomAD | |
| rs1775971228 | 400 | V>A | No | TOPMed | |
| rs2127944796 | 401 | T>A | No | Ensembl | |
| rs751791155 | 404 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1270412615 | 406 | L>P | No | gnomAD | |
| TCGA novel | 407 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1775970535 | 407 | A>T | No | Ensembl | |
| rs1368174232 | 408 | H>N | No |
TOPMed gnomAD |
|
|
COSM1082669 rs145928664 |
408 | H>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1418489484 | 409 | A>G | No | gnomAD | |
| rs1402331883 | 409 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1402331883 | 409 | A>T | No |
TOPMed gnomAD |
|
| TCGA novel | 410 | N>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4847703 COSM4847702 |
416 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1397320389 | 418 | V>L | No |
TOPMed gnomAD |
|
| rs1327337269 | 419 | N>S | No | gnomAD | |
| rs1775968290 | 421 | V>I | No | gnomAD | |
|
COSM4393164 COSM4393165 |
422 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1169903293 | 423 | D>G | No | gnomAD | |
|
rs1775967400 COSM48865 |
426 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs542602078 | 426 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs542602078 | 426 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs542602078 | 426 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs866023703 | 427 | S>F | No | Ensembl | |
| rs149093618 | 428 | Q>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs771888219 | 429 | R>K | No |
ExAC gnomAD |
|
| rs1179484555 | 430 | L>F | No |
TOPMed gnomAD |
|
| rs199839585 | 430 | L>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1582585159 | 431 | F>V | No | Ensembl | |
| rs1273384862 | 434 | V>I | No | gnomAD | |
| rs144991363 | 435 | S>C | No |
ESP ExAC gnomAD |
|
| rs144991363 | 435 | S>G | No |
ESP ExAC gnomAD |
|
| rs749253547 | 436 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1234366311 | 439 | G>S | No | gnomAD | |
|
COSM6175035 COSM6175036 |
440 | Q>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 441 | A>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781072828 | 441 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1378155962 | 442 | A>P | No | TOPMed | |
| rs773950345 | 444 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs749130416 | 445 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs1562059593 | 448 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs769773179 | 448 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1771271575 | 449 | V>G | No | Ensembl | |
|
rs1400112416 COSM3922257 COSM3922256 |
450 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs754573060 | 450 | M>R | No | Ensembl | |
| rs754573060 | 450 | M>T | No | Ensembl | |
| rs758549619 | 451 | K>E | No |
ExAC gnomAD |
|
| rs201935163 | 452 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1164322567 | 452 | E>G | No | TOPMed | |
| rs147680635 | 453 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 453 | R>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs779416238 | 457 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs779416238 | 457 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1484017296 | 457 | R>W | No | gnomAD | |
| rs1771268324 | 458 | S>N | No | Ensembl | |
| rs1771268491 | 458 | S>R | No | Ensembl | |
| rs191102962 | 459 | V>A | No | 1000Genomes | |
| TCGA novel | 460 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM4357488 COSM1205479 rs1448138791 |
460 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs555480655 | 462 | S>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM4357487 COSM3178863 |
462 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1231930557 | 463 | W>R | No | gnomAD | |
| rs1334631229 | 465 | E>K | No | gnomAD | |
| rs1294264997 | 466 | P>Q | No | gnomAD | |
| TCGA novel | 466 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM6175038 COSM6175037 |
467 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1332501890 | 467 | E>K | No |
TOPMed gnomAD |
|
| rs1332501890 | 467 | E>Q | No |
TOPMed gnomAD |
|
| rs1771266349 | 467 | E>V | No | TOPMed | |
| rs774074768 | 469 | P>R | No |
ExAC gnomAD |
|
| rs1771265834 | 470 | N>S | No | gnomAD | |
| rs1232168679 | 472 | V>A | No | TOPMed | |
| rs1431588483 | 473 | I>V | No |
TOPMed gnomAD |
|
| rs1771265031 | 474 | T>A | No | gnomAD | |
| rs762787694 | 475 | E>* | No |
ExAC gnomAD |
|
| TCGA novel | 475 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762787694 | 475 | E>K | No |
ExAC gnomAD |
|
| rs762787694 | 475 | E>Q | No |
ExAC gnomAD |
|
| rs1459766148 | 476 | Y>C | No | gnomAD | |
| rs1165179670 | 477 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1771264182 | 478 | I>L | No | Ensembl | |
| rs369681580 | 480 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| COSM70627 | 482 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs867625037 COSM4895548 COSM4895549 |
482 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs867625037 | 482 | E>Q | No |
TOPMed gnomAD |
|
| rs371905556 | 485 | Q>E | No | ESP | |
| rs1771113438 | 487 | E>D | No |
TOPMed gnomAD |
|
| rs776583281 | 487 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1289972813 | 487 | E>V | No |
TOPMed gnomAD |
|
| rs968186662 | 488 | R>P | No |
TOPMed gnomAD |
|
| rs968186662 | 488 | R>Q | No |
TOPMed gnomAD |
|
| rs139833485 | 488 | R>W | No |
1000Genomes ESP ExAC gnomAD |
|
| rs1371749537 | 489 | T>I | No | gnomAD | |
|
COSM3831247 COSM3831248 |
491 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127877662 | 492 | T>I | No | Ensembl | |
| rs1388409740 | 493 | V>L | No | gnomAD | |
|
COSM5011647 COSM5011646 |
494 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1771112023 | 498 | T>S | No | Ensembl | |
| rs1277605416 | 500 | A>T | No |
TOPMed gnomAD |
|
|
COSM3630613 rs749473870 COSM4891847 |
501 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1372005745 | 502 | I>V | No | gnomAD | |
| rs267601170 | 504 | N>I | No | Ensembl | |
| rs1771110726 | 504 | N>K | No | Ensembl | |
| rs1771110518 | 507 | P>A | No | Ensembl | |
| rs1771110518 | 507 | P>S | No | Ensembl | |
|
COSM4893666 rs2127877576 COSM3630612 |
508 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127877583 | 508 | G>R | No | Ensembl | |
| rs778224828 | 510 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs778224828 | 510 | V>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 510 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 511 | Y>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 512 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 513 | F>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1377657771 | 514 | Q>H | No | gnomAD | |
| rs756567371 | 514 | Q>R | No |
ExAC gnomAD |
|
| rs1178898166 | 515 | I>V | No |
TOPMed gnomAD |
|
| rs2127877543 | 516 | R>Q | No | 1000Genomes | |
|
COSM4979814 COSM4979815 |
516 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1264422608 | 518 | F>V | No | gnomAD | |
| rs986768697 | 520 | A>D | No | Ensembl | |
| rs1771109152 | 520 | A>T | No | Ensembl | |
| rs1417829621 | 521 | A>S | No | gnomAD | |
| rs954614070 | 523 | Y>H | No |
TOPMed gnomAD |
|
| rs954614070 | 523 | Y>N | No |
TOPMed gnomAD |
|
| rs748625367 | 524 | G>A | No |
ExAC gnomAD |
|
| rs569672013 | 525 | N>H | No |
1000Genomes ExAC gnomAD |
|
| rs369029096 | 526 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1771107783 | 527 | S>G | No | Ensembl | |
| COSM280831 | 527 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1771107622 | 527 | S>T | No | TOPMed | |
| rs1348732539 | 529 | R>G | No |
TOPMed gnomAD |
|
| rs1288102655 | 530 | L>H | No | TOPMed | |
| rs1771106437 | 533 | A>P | No | Ensembl | |
| TCGA novel | 535 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1009626356 COSM3922255 COSM232257 |
537 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1771105192 | 542 | M>I | No | TOPMed | |
| rs1771105435 | 542 | M>K | No | TOPMed | |
| rs1355331916 | 543 | F>L | No | gnomAD | |
| rs1000520166 | 546 | T>I | No | Ensembl | |
| rs1380030438 | 547 | A>D | No | gnomAD | |
| rs148781580 | 547 | A>P | No |
ESP gnomAD |
|
| rs148781580 | 547 | A>T | No |
ESP gnomAD |
|
|
rs1380030438 COSM4992797 COSM4992796 |
547 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs934498317 | 550 | S>G | No | Ensembl | |
| rs1770846171 | 550 | S>I | No | TOPMed | |
| rs928479836 | 552 | Q>E | No | Ensembl | |
| rs1459246528 | 552 | Q>H | No |
TOPMed gnomAD |
|
| rs756964921 | 552 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1164650086 | 555 | V>A | No | gnomAD | |
| rs753621830 | 559 | A>V | No | ExAC | |
| rs1770844479 | 560 | V>M | No | TOPMed | |
| rs1770844177 | 561 | V>A | No | TOPMed | |
|
COSM6107507 COSM6107506 |
565 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM70626 rs77295523 |
566 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1428020526 | 567 | I>M | No | gnomAD | |
| rs760600822 | 567 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1770843504 | 568 | I>V | No | TOPMed | |
| rs773473690 | 570 | V>A | No |
ExAC gnomAD |
|
| rs762911058 | 570 | V>L | No |
ExAC gnomAD |
|
| rs762911058 | 570 | V>M | No |
ExAC gnomAD |
|
| rs770136433 | 572 | M>L | No |
ExAC gnomAD |
|
| TCGA novel | 572 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1770842692 | 573 | V>A | No | Ensembl | |
| TCGA novel | 575 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs546509392 | 576 | F>V | No | Ensembl | |
| rs1562053469 | 577 | I>V | No | Ensembl | |
| rs1391451187 | 578 | I>T | No |
TOPMed gnomAD |
|
| rs1377724860 | 578 | I>V | No |
TOPMed gnomAD |
|
| rs1770841488 | 579 | G>E | No | TOPMed | |
| rs1356700106 | 579 | G>W | No | gnomAD | |
| rs1041561838 | 580 | R>* | No | TOPMed | |
|
COSM1082665 COSM4866646 |
581 | R>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777573184 | 583 | C>G | No |
ExAC gnomAD |
|
| rs755890860 | 584 | G>V | No |
ExAC gnomAD |
|
| rs1036447486 | 586 | S>R | No |
TOPMed gnomAD |
|
| rs1432954439 | 587 | K>E | No | gnomAD | |
| rs1770806253 | 589 | D>G | No |
TOPMed gnomAD |
|
|
COSM3831246 COSM3831245 |
589 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1582410223 | 590 | Q>H | No | TOPMed | |
| rs767334617 | 591 | E>G | No |
ExAC gnomAD |
|
| rs750462795 | 593 | D>N | No |
ExAC gnomAD |
|
| rs1483753655 | 595 | E>* | No | gnomAD | |
| rs1483753655 | 595 | E>Q | No | gnomAD | |
| rs1770805076 | 597 | Y>H | No | TOPMed | |
|
COSM4396303 COSM4396304 rs1179394255 |
599 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2127860659 | 600 | F>C | No | Ensembl | |
| TCGA novel | 601 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1251347 | 601 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748898049 | 602 | F>L | No |
ExAC TOPMed gnomAD |
|
|
COSM3876205 COSM3876206 |
602 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4402213 COSM3630609 rs868510824 |
603 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs777258745 | 605 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1236447200 | 607 | T>A | No | gnomAD | |
| rs755839039 | 609 | I>T | No |
ExAC gnomAD |
|
| TCGA novel | 609 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376168086 | 613 | T>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1282784862 | 613 | T>P | No |
TOPMed gnomAD |
|
| rs1330278383 | 614 | Y>C | No | gnomAD | |
|
COSM3995163 COSM3995162 |
615 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1770589860 | 615 | E>K | No | Ensembl | |
| rs1562049937 | 616 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs373264389 | 617 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs373264389 | 617 | P>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4785646 COSM1446489 |
618 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs946160133 | 618 | N>K | No | TOPMed | |
| rs751571551 | 619 | R>G | No |
ExAC gnomAD |
|
| rs1770588980 | 620 | A>S | No |
TOPMed gnomAD |
|
| rs1487499869 | 621 | V>A | No | TOPMed | |
| rs200277966 | 622 | H>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs200277966 | 622 | H>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1770588510 | 623 | Q>K | No | Ensembl | |
| rs754002431 | 623 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs149602525 | 625 | A>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs149602525 | 625 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM6175040 COSM6175039 |
626 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1160371803 | 629 | D>G | No | TOPMed | |
|
COSM4813037 COSM451944 |
629 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1412626348 | 630 | A>D | No | gnomAD | |
| rs139211112 | 631 | S>T | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 632 | C>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1770586775 | 634 | K>I | No | TOPMed | |
| rs1239820687 | 634 | K>N | No | gnomAD | |
| rs1770586549 | 635 | I>T | No | TOPMed | |
| rs1188528023 | 636 | E>K | No | gnomAD | |
| rs1270476760 | 637 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs774868013 | 637 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1360471715 | 638 | V>M | No | TOPMed | |
| rs771465590 | 639 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1770585746 | 639 | I>V | No | Ensembl | |
| rs531906513 | 640 | G>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs531906513 | 640 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs992785889 | 641 | A>S | No |
TOPMed gnomAD |
|
| rs992785889 | 641 | A>T | No |
TOPMed gnomAD |
|
|
rs766856886 COSM1697648 COSM5608521 |
642 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2127859217 | 643 | E>A | No | Ensembl | |
| rs367646826 | 645 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs367646826 | 645 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1582401525 | 645 | G>V | No | Ensembl | |
| rs1019151131 | 646 | E>A | No | TOPMed | |
| rs534228190 | 646 | E>K | No | 1000Genomes | |
| rs769370190 | 648 | C>F | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 649 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1193284113 | 649 | S>R | No | gnomAD | |
| rs1770530827 | 650 | G>D | No | TOPMed | |
| rs571637670 | 651 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs776394413 | 651 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs776394413 | 651 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs571637670 | 651 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1206076675 | 652 | L>F | No |
TOPMed gnomAD |
|
| rs1484095270 | 653 | K>I | No | gnomAD | |
| rs768474651 | 655 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1316144079 | 655 | P>R | No |
TOPMed gnomAD |
|
|
COSM1082660 COSM4870279 rs768474651 |
655 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
|
COSM3922254 COSM3922253 |
656 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1231037263 | 656 | G>R | No | gnomAD | |
| COSM98136 | 656 | G>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1277577151 | 657 | K>R | No |
TOPMed gnomAD |
|
| rs1311518360 | 658 | R>K | No |
TOPMed gnomAD |
|
| rs1311518360 | 658 | R>T | No |
TOPMed gnomAD |
|
|
COSM4782548 COSM3749989 |
659 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs780064217 | 660 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs780064217 | 660 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs758348899 | 661 | A>S | No | ExAC | |
| rs745827385 | 661 | A>V | No |
ExAC gnomAD |
|
| rs202223312 | 662 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs202223312 | 662 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs953493697 | 663 | A>T | No | TOPMed | |
| rs1770527799 | 663 | A>V | No | Ensembl | |
| rs1263935491 | 664 | I>L | No |
TOPMed gnomAD |
|
| rs1770527266 | 664 | I>M | No | TOPMed | |
| rs1300555214 | 664 | I>T | No |
TOPMed gnomAD |
|
| rs767702926 | 666 | T>I | No |
ExAC gnomAD |
|
| rs2127859101 | 667 | L>M | No | Ensembl | |
| rs1415493097 | 668 | K>Q | No | gnomAD | |
| rs766734179 | 669 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs751915816 | 669 | V>L | No |
ExAC gnomAD |
|
|
COSM6175042 COSM6175041 |
670 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1458314724 | 671 | Y>* | No |
TOPMed gnomAD |
|
| rs1206527280 | 672 | T>A | No | TOPMed | |
| rs1231158173 | 674 | K>E | No | Ensembl | |
| rs201814256 | 675 | Q>K | No |
ExAC TOPMed gnomAD |
|
| rs146203229 | 675 | Q>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373761211 | 676 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs760411632 | 676 | R>S | No |
ExAC gnomAD |
|
| rs373761211 | 676 | R>T | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 677 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1770524764 | 678 | D>E | No | TOPMed | |
|
COSM4897388 rs1256527658 COSM3630606 |
678 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM1082658 COSM4872453 |
679 | F>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1770524334 | 680 | L>V | No |
TOPMed gnomAD |
|
| rs771918424 | 681 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1228694029 | 684 | S>G | No | gnomAD | |
| rs1353290090 | 684 | S>I | No | TOPMed | |
| rs1228694029 | 684 | S>R | No | gnomAD | |
| rs1770523323 | 685 | I>V | No | TOPMed | |
| rs1770523043 | 686 | M>R | No | Ensembl | |
| rs1770523188 | 686 | M>V | No | TOPMed | |
| rs1284327490 | 690 | D>G | No | TOPMed | |
| rs778876357 | 692 | P>A | No |
ExAC gnomAD |
|
| rs770820190 | 692 | P>L | No |
ExAC gnomAD |
|
| rs1770521941 | 693 | N>S | No |
TOPMed gnomAD |
|
| rs569112388 | 694 | V>I | No |
1000Genomes ExAC gnomAD |
|
|
COSM597707 COSM4871270 |
696 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1045593838 | 696 | H>P | No | TOPMed | |
| rs1045593838 | 696 | H>R | No | TOPMed | |
| rs1770521274 | 698 | E>A | No | Ensembl | |
|
COSM1697647 rs138518092 COSM4893650 |
698 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1388428555 | 700 | V>D | No | gnomAD | |
| rs1770521024 | 700 | V>I | No | TOPMed | |
| TCGA novel | 701 | V>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1770520511 | 702 | T>A | No | Ensembl | |
| rs1423543958 | 702 | T>K | No | gnomAD | |
| rs370400794 | 703 | R>I | No | Ensembl | |
|
COSM4897516 COSM3630605 |
704 | G>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749228043 | 706 | P>L | No |
ExAC gnomAD |
|
|
rs749228043 TCGA novel |
706 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
| rs1008162814 | 706 | P>S | No | Ensembl | |
| rs1247358846 | 708 | M>I | No | gnomAD | |
| TCGA novel | 708 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1770491622 | 708 | M>V | No | Ensembl | |
| rs889594472 | 709 | I>L | No |
TOPMed gnomAD |
|
| rs1307419633 | 709 | I>T | No |
TOPMed gnomAD |
|
| rs1365931416 | 710 | V>A | No | TOPMed | |
| rs781190788 | 711 | I>T | No |
ExAC gnomAD |
|
| rs1406597161 | 712 | E>G | No | TOPMed | |
| rs747133463 | 713 | F>I | No |
ExAC gnomAD |
|
| rs747133463 | 713 | F>L | No |
ExAC gnomAD |
|
| rs1208917339 | 714 | M>I | No | gnomAD | |
| rs1050029945 | 714 | M>L | No | Ensembl | |
| rs370612964 | 714 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs150176645 | 715 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1770490108 | 718 | A>V | No | TOPMed | |
| rs376794821 | 720 | D>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs983485848 | 720 | D>G | No |
TOPMed gnomAD |
|
| rs1770489462 | 721 | A>S | No | Ensembl | |
| rs1770489358 | 721 | A>V | No | Ensembl | |
|
COSM3876204 COSM3876203 |
724 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4897255 COSM3630603 |
724 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs775497224 | 726 | H>R | No |
ExAC gnomAD |
|
|
COSM4893609 COSM3630602 |
727 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746088117 | 728 | G>R | No |
ExAC TOPMed gnomAD |
|
|
COSM4810982 rs1270091812 COSM1312457 |
731 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs1770427495 | 733 | I>V | No | Ensembl | |
| rs749714596 | 734 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1327213322 | 737 | G>E | No | gnomAD | |
|
rs1208009700 COSM48342 |
738 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1770426537 | 738 | M>T | No | Ensembl | |
| rs752253692 | 741 | G>R | No |
ExAC gnomAD |
|
| rs1770425887 | 742 | I>T | No | Ensembl | |
| rs1770426017 | 742 | I>V | No | gnomAD | |
| TCGA novel | 743 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1243702457 | 744 | A>G | No | TOPMed | |
| rs1770425622 | 744 | A>T | No | TOPMed | |
| rs1484087506 | 746 | M>I | No |
TOPMed gnomAD |
|
| rs183610251 | 748 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs1770424725 | 748 | Y>H | No | TOPMed | |
| rs1405423810 | 750 | A>T | No | gnomAD | |
| rs2127856626 | 751 | D>V | No | Ensembl | |
| rs1419285880 | 752 | M>V | No | TOPMed | |
|
COSM4903784 COSM4903785 |
753 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1392330402 | 753 | G>R | No |
TOPMed gnomAD |
|
| rs1770423590 | 753 | G>V | No | Ensembl | |
| rs751194338 | 757 | R>K | No |
ExAC TOPMed gnomAD |
|
| rs1770423314 | 758 | D>N | No | TOPMed | |
|
COSM3630599 COSM4905524 |
762 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs762713500 | 762 | R>H | No |
ExAC gnomAD |
|
| rs1770422912 | 763 | N>D | No | Ensembl | |
| rs1472053844 | 763 | N>S | No | gnomAD | |
| rs1770422508 | 764 | I>N | No | TOPMed | |
| rs773047453 | 764 | I>V | No |
ExAC gnomAD |
|
| rs1440140016 | 769 | N>S | No |
TOPMed gnomAD |
|
| rs964339668 | 770 | L>F | No | TOPMed | |
| rs746058123 | 771 | V>I | No |
ExAC gnomAD |
|
| rs771331966 | 772 | C>G | No |
ExAC TOPMed gnomAD |
|
| rs1281423342 | 773 | K>E | No |
TOPMed gnomAD |
|
| rs141289390 | 778 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1408104586 | 780 | S>P | No | gnomAD | |
| rs1327105282 | 781 | R>* | No |
TOPMed gnomAD |
|
| rs372024057 | 781 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs372024057 | 781 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1178526425 | 783 | I>T | No | gnomAD | |
| rs751128749 | 783 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1770418775 | 785 | D>G | No | Ensembl | |
| rs1770418490 | 787 | P>S | No | gnomAD | |
| TCGA novel | 788 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127856499 | 789 | A>V | No | Ensembl | |
| rs1770417891 | 791 | Y>* | No | Ensembl | |
| rs758111523 | 791 | Y>C | No |
ExAC gnomAD |
|
| rs1214294974 | 791 | Y>D | No | gnomAD | |
| rs758111523 | 791 | Y>F | No |
ExAC gnomAD |
|
| TCGA novel | 791 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1562047011 | 792 | T>K | No | gnomAD | |
| rs1644838588 | 792 | T>S | No | TOPMed | |
| rs1770417513 | 794 | T>A | No | TOPMed | |
| rs1770417513 | 794 | T>P | No | TOPMed | |
| rs773655407 | 795 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs773655407 | 795 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1435219791 | 796 | G>E | No | Ensembl | |
|
COSM4537152 COSM4537153 |
796 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs770305987 | 798 | I>L | No |
ExAC gnomAD |
|
|
COSM6107510 COSM6107511 |
800 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1582395836 | 801 | R>G | No | Ensembl | |
| rs1770362959 | 801 | R>K | No | Ensembl | |
|
COSM4167323 COSM4167324 |
801 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs191032715 | 802 | W>R | No | 1000Genomes | |
|
COSM3922249 COSM3922250 rs1219702649 |
804 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1770362341 | 805 | P>H | No | TOPMed | |
|
rs775779069 COSM4758893 COSM4758892 |
806 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs746495128 | 807 | A>S | No |
ExAC gnomAD |
|
|
COSM3928662 COSM3928661 |
808 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4856998 COSM484459 |
809 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs757986575 | 809 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs757986575 | 809 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs147054938 | 811 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs147054938 | 811 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1295357534 | 811 | R>W | No | gnomAD | |
| rs940477878 | 812 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
|
COSM3630598 COSM4904037 |
814 | T>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs778636182 | 815 | S>P | No |
ExAC gnomAD |
|
| rs145700620 | 816 | A>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs1770360912 | 816 | A>S | No | Ensembl | |
| rs145700620 | 816 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1433933397 | 818 | D>V | No |
TOPMed gnomAD |
|
| rs1348945329 | 819 | V>A | No | gnomAD | |
| rs1770360432 | 819 | V>I | No | Ensembl | |
|
COSM3630597 COSM4897408 |
820 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs199833723 | 821 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs1275298374 | 821 | S>N | No | TOPMed | |
| TCGA novel | 821 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760735153 | 822 | Y>C | No |
ExAC gnomAD |
|
| TCGA novel | 822 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200484495 | 824 | I>V | No | 1000Genomes | |
| rs766582260 | 826 | M>L | No | ExAC | |
| rs1422163923 | 826 | M>T | No | gnomAD | |
| rs766582260 | 826 | M>V | No | ExAC | |
| TCGA novel | 827 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 827 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs867402067 | 828 | E>K | No | Ensembl | |
| rs1770358334 | 829 | V>A | No | TOPMed | |
| rs1770358472 | 829 | V>I | No | Ensembl | |
| rs763219152 | 830 | M>T | No |
ExAC gnomAD |
|
| rs773600194 | 832 | Y>C | No |
ExAC gnomAD |
|
|
rs568334270 COSM4904231 COSM3630596 |
833 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1051471746 | 833 | G>R | No | Ensembl | |
| rs568334270 | 833 | G>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM6107513 COSM6107512 |
834 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1304791200 | 836 | P>T | No | gnomAD | |
| rs1770357260 | 837 | Y>C | No |
TOPMed gnomAD |
|
| rs1273984232 | 838 | W>R | No | gnomAD | |
| rs1342659278 | 843 | Q>E | No |
TOPMed gnomAD |
|
|
COSM4859319 COSM743818 |
844 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777118725 | 844 | D>V | No |
ExAC gnomAD |
|
| rs2127847329 | 845 | V>D | No | Ensembl | |
| rs1320953732 | 845 | V>F | No | gnomAD | |
| rs1320953732 | 845 | V>I | No | gnomAD | |
| rs1320953732 | 845 | V>L | No | gnomAD | |
| rs538121970 | 846 | I>T | No | Ensembl | |
| rs2127847321 | 847 | K>* | No | Ensembl | |
| rs2127847318 | 847 | K>I | No | Ensembl | |
| rs1347209840 | 847 | K>N | No | Ensembl | |
| rs2127847318 | 847 | K>T | No | Ensembl | |
| rs2127847310 | 848 | A>E | No | Ensembl | |
| rs2127847310 | 848 | A>G | No | Ensembl | |
| rs2127847310 | 848 | A>V | No | Ensembl | |
| rs2127847299 | 849 | I>K | No | Ensembl | |
| rs762098670 | 849 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs2127847299 | 849 | I>R | No | Ensembl | |
| rs2127847299 | 849 | I>T | No | Ensembl | |
| rs762098670 | 849 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2127847292 | 850 | E>D | No | Ensembl | |
| rs1038362394 | 850 | E>K | No | Ensembl | |
|
COSM4861971 rs1038362394 COSM743819 |
850 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127847294 | 850 | E>V | No | Ensembl | |
| TCGA novel | 851 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127847286 | 851 | E>D | No | Ensembl | |
| rs2127847290 | 851 | E>K | No | Ensembl | |
| rs2127847290 | 851 | E>Q | No | Ensembl | |
| rs2127847288 | 851 | E>V | No | Ensembl | |
| rs1287261842 | 852 | G>A | No | gnomAD | |
| rs2127847284 | 852 | G>C | No | Ensembl | |
|
COSM3876202 rs1287261842 COSM3876201 |
852 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs2127847284 | 852 | G>R | No | Ensembl | |
| rs2127847284 | 852 | G>S | No | Ensembl | |
| rs1301647844 | 853 | Y>* | No | gnomAD | |
| rs1374830452 | 853 | Y>C | No | gnomAD | |
|
COSM743820 rs1465960814 COSM4481974 |
854 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM3876199 COSM3876200 rs776802108 |
854 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs776802108 | 854 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs776802108 | 854 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs2127847254 | 855 | L>* | No | Ensembl | |
| rs2127847257 | 855 | L>I | No | Ensembl | |
| rs2127847248 | 856 | P>A | No | Ensembl | |
| rs2127847244 | 856 | P>L | No | Ensembl | |
| rs2127847244 | 856 | P>Q | No | Ensembl | |
| rs2127847244 | 856 | P>R | No | Ensembl | |
| rs2127847248 | 856 | P>S | No | Ensembl | |
| rs2127847248 | 856 | P>T | No | Ensembl | |
| rs2127847236 | 857 | A>G | No | Ensembl | |
| rs2127847239 | 857 | A>P | No | Ensembl | |
| rs2127847239 | 857 | A>S | No | Ensembl | |
| rs2127847239 | 857 | A>T | No | Ensembl | |
| rs2127847236 | 857 | A>V | No | Ensembl | |
| rs2127847227 | 858 | P>A | No | Ensembl | |
| rs2127847224 | 858 | P>H | No | Ensembl | |
| rs2127847224 | 858 | P>L | No | Ensembl | |
| rs2127847224 | 858 | P>R | No | Ensembl | |
|
COSM3394355 rs2127847227 COSM4945846 |
858 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127847219 | 859 | M>I | No | Ensembl | |
| rs200816651 | 859 | M>K | No |
TOPMed gnomAD |
|
| rs1179028901 | 859 | M>V | No |
TOPMed gnomAD |
|
| rs113927523 | 860 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2127847215 | 860 | D>H | No | Ensembl | |
| rs2127847215 | 860 | D>N | No | Ensembl | |
| rs2127847215 | 860 | D>Y | No | Ensembl | |
| rs2127847201 | 861 | C>F | No | Ensembl | |
| rs1393547166 | 861 | C>R | No | Ensembl | |
| rs2127847201 | 861 | C>S | No | Ensembl | |
| rs1393547166 | 861 | C>S | No | Ensembl | |
| rs2127847198 | 861 | C>W | No | Ensembl | |
| rs2127847201 | 861 | C>Y | No | Ensembl | |
| rs2127847194 | 862 | P>A | No | Ensembl | |
| rs2127847189 | 862 | P>L | No | Ensembl | |
| rs2127847189 | 862 | P>Q | No | Ensembl | |
| rs2127847189 | 862 | P>R | No | Ensembl | |
| rs2127847194 | 862 | P>S | No | Ensembl | |
| rs2127847194 | 862 | P>T | No | Ensembl | |
| rs2127847180 | 863 | A>G | No | Ensembl | |
| rs377619656 | 863 | A>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs377619656 | 863 | A>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs377619656 | 863 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2127847180 | 863 | A>V | No | Ensembl | |
| rs2127847171 | 864 | G>A | No | Ensembl | |
| rs2127847171 | 864 | G>D | No | Ensembl | |
| rs2127847171 | 864 | G>V | No | Ensembl | |
| rs2127847163 | 865 | L>H | No | Ensembl | |
| rs2127847167 | 865 | L>I | No | Ensembl | |
| rs2127847163 | 865 | L>P | No | Ensembl | |
| rs2127847167 | 865 | L>V | No | Ensembl | |
| rs2127847155 | 866 | H>L | No | Ensembl | |
| rs2127847155 | 866 | H>P | No | Ensembl | |
| rs2127847150 | 866 | H>Q | No | Ensembl | |
|
COSM3630594 COSM4897173 |
866 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2127847148 | 867 | Q>* | No | Ensembl | |
| rs2127847148 | 867 | Q>E | No | Ensembl | |
| rs1254132234 | 867 | Q>H | No | gnomAD | |
| rs2127847138 | 868 | L>Q | No | Ensembl | |
| rs2127847142 | 868 | L>V | No | Ensembl | |
| rs1769979835 | 869 | M>I | No | Ensembl | |
| TCGA novel | 869 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127847122 | 870 | L>* | No | Ensembl | |
| rs2127847116 | 870 | L>F | No | Ensembl | |
| rs2127847126 | 870 | L>M | No | Ensembl | |
|
rs2127847112 COSM3411342 COSM4782122 |
871 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127847112 | 871 | D>N | No | Ensembl | |
| rs2127847110 | 871 | D>V | No | Ensembl | |
| rs2127847112 | 871 | D>Y | No | Ensembl | |
| rs2127847106 | 872 | C>* | No | Ensembl | |
| rs2127847109 | 872 | C>S | No | Ensembl | |
| rs1266282599 | 873 | W>* | No |
TOPMed gnomAD |
|
| rs2127847099 | 873 | W>* | No | Ensembl | |
| rs1266282599 | 873 | W>C | No |
TOPMed gnomAD |
|
|
rs2127847099 COSM3630593 COSM4903912 |
873 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127847102 | 873 | W>R | No | Ensembl | |
| rs2127847099 | 873 | W>S | No | Ensembl | |
| rs1769979537 | 874 | Q>* | No | gnomAD | |
| rs1769979537 | 874 | Q>E | No | gnomAD | |
| rs745373107 | 874 | Q>P | No |
ExAC gnomAD |
|
| rs374483308 | 875 | K>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs2127847094 | 875 | K>R | No | Ensembl | |
| rs2127847084 | 876 | E>D | No | Ensembl | |
| rs2127847085 | 876 | E>G | No | Ensembl | |
| rs748985745 | 876 | E>K | No |
ExAC gnomAD |
|
| rs748985745 | 876 | E>Q | No |
ExAC gnomAD |
|
| rs2127847085 | 876 | E>V | No | Ensembl | |
| rs777383467 | 877 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs777383467 | 877 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM4787387 COSM3765301 rs755970343 |
877 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs755970343 COSM6107514 COSM6107515 |
877 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs755970343 | 877 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs777383467 | 877 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs923098531 | 878 | A>G | No |
TOPMed gnomAD |
|
| rs1769978146 | 878 | A>P | No | gnomAD | |
| rs1769978146 | 878 | A>S | No | gnomAD | |
| rs1769978146 | 878 | A>T | No | gnomAD | |
| rs923098531 | 878 | A>V | No |
TOPMed gnomAD |
|
| rs2127847062 | 879 | E>* | No | Ensembl | |
| rs1309944126 | 879 | E>A | No | gnomAD | |
|
rs2127847062 COSM6107516 COSM6107517 |
879 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127847062 | 879 | E>Q | No | Ensembl | |
| rs2127847056 | 880 | R>G | No | Ensembl | |
| rs1769977744 | 880 | R>K | No | Ensembl | |
| rs2127847048 | 880 | R>S | No | Ensembl | |
| rs2127847043 | 881 | P>A | No | Ensembl | |
| rs2127847043 | 881 | P>S | No | Ensembl | |
| rs2127847043 | 881 | P>T | No | Ensembl | |
| rs2127847039 | 882 | K>* | No | Ensembl | |
| rs1769977601 | 882 | K>I | No | TOPMed | |
| rs747995954 | 882 | K>N | No |
ExAC gnomAD |
|
| rs1769977601 | 882 | K>T | No | TOPMed | |
| rs2127847030 | 883 | F>Y | No | Ensembl | |
| rs781260960 | 884 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2127847028 | 884 | E>K | No | Ensembl | |
| rs2127847028 | 884 | E>Q | No | Ensembl | |
| rs2127847025 | 884 | E>V | No | Ensembl | |
| rs2127847014 | 885 | Q>H | No | Ensembl | |
| rs2127847022 | 885 | Q>K | No | Ensembl | |
| rs1769977138 | 885 | Q>R | No | TOPMed | |
| rs2127847010 | 886 | I>K | No | Ensembl | |
| rs755032746 | 886 | I>L | No |
ExAC gnomAD |
|
| rs2127847008 | 886 | I>M | No | Ensembl | |
| rs2127847010 | 886 | I>T | No | Ensembl | |
| rs2127847002 | 887 | V>D | No | Ensembl | |
| rs2127847006 | 887 | V>F | No | Ensembl | |
| rs2127847006 | 887 | V>L | No | Ensembl | |
| rs2127846993 | 888 | G>* | No | Ensembl | |
| rs750460575 | 888 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs2127846993 | 888 | G>R | No | Ensembl | |
| rs750460575 | 888 | G>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 888 | G>W | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2127846983 | 889 | I>L | No | Ensembl | |
| rs1769976745 | 889 | I>S | No | gnomAD | |
| rs1769976745 | 889 | I>T | No | gnomAD | |
| rs2127846983 | 889 | I>V | No | Ensembl | |
| rs1769976642 | 890 | L>I | No | TOPMed | |
| rs2127846970 | 890 | L>Q | No | Ensembl | |
|
COSM6107518 COSM6107519 rs1769976642 |
890 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs2127846959 | 891 | D>E | No | Ensembl | |
| rs765371643 | 891 | D>G | No |
ExAC gnomAD |
|
| rs2127846963 | 891 | D>H | No | Ensembl | |
| rs2127846963 | 891 | D>N | No | Ensembl | |
| rs765371643 | 891 | D>V | No |
ExAC gnomAD |
|
|
COSM6107520 rs2127846963 COSM6107521 |
891 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1769976405 | 892 | K>Q | No | TOPMed | |
| rs2127846956 | 892 | K>R | No | Ensembl | |
|
COSM4905810 rs2127846947 COSM3630591 |
893 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA Cosmic |
| rs757369957 | 893 | M>K | No |
ExAC TOPMed gnomAD |
|
| rs2127846951 | 893 | M>L | No | Ensembl | |
| rs757369957 | 893 | M>R | No |
ExAC TOPMed gnomAD |
|
|
COSM6175043 COSM6175044 rs757369957 |
893 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC TOPMed gnomAD |
| rs2127846945 | 894 | I>F | No | Ensembl | |
| rs2127846945 | 894 | I>L | No | Ensembl | |
|
COSM167563 rs1035177735 COSM4868727 |
895 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM4814395 COSM451943 rs1035177735 |
895 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs754119070 | 895 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs754119070 | 895 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM1446484 COSM4948483 rs754119070 |
895 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2127846932 | 896 | N>I | No | Ensembl | |
| rs764259302 | 896 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs2127846935 | 896 | N>Y | No | Ensembl | |
| rs2127846924 | 897 | P>A | No | Ensembl | |
| rs2127846919 | 897 | P>L | No | Ensembl | |
| rs2127846919 | 897 | P>Q | No | Ensembl | |
| rs2127846919 | 897 | P>R | No | Ensembl | |
| rs2127846924 | 897 | P>S | No | Ensembl | |
| rs2127846924 | 897 | P>T | No | Ensembl | |
| rs1769975532 | 898 | N>D | No | Ensembl | |
| rs2127846914 | 898 | N>I | No | Ensembl | |
| rs2127846913 | 898 | N>K | No | Ensembl | |
| rs1445240842 | 899 | S>C | No | gnomAD | |
| rs1445240842 | 899 | S>G | No | gnomAD | |
|
TCGA novel rs2127846905 |
899 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs2127846904 | 899 | S>R | No | Ensembl | |
| rs2127846905 | 899 | S>T | No | Ensembl | |
| rs2127846896 | 900 | L>P | No | Ensembl | |
| rs2127846896 | 900 | L>Q | No | Ensembl | |
| rs2127846900 | 900 | L>V | No | Ensembl | |
| rs2127846890 | 901 | K>N | No | Ensembl | |
|
rs775894675 COSM3928015 |
902 | T>I | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs2127846887 | 902 | T>P | No | Ensembl | |
| rs775894675 | 902 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs2127846887 | 902 | T>S | No | Ensembl | |
| rs2127846874 | 903 | P>A | No | Ensembl | |
| rs768012065 | 903 | P>H | No |
ExAC gnomAD |
|
|
COSM3630590 COSM4897498 rs768012065 |
903 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC gnomAD |
| rs768012065 | 903 | P>R | No |
ExAC gnomAD |
|
|
VAR_042152 COSM21146 rs2127846874 COSM4357441 |
903 | P>S | Variant assessed as Somatic; MODERATE impact. a metastatic melanoma sample; somatic mutation [NCI-TCGA, UniProt] | No |
NCI-TCGA Cosmic Ensembl UniProt |
| rs2127846874 | 903 | P>T | No | Ensembl | |
| rs770497783 | 904 | L>P | No |
ExAC gnomAD |
|
| rs770497783 | 904 | L>Q | No |
ExAC gnomAD |
|
| rs773680518 | 904 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2127846854 | 905 | G>A | No | Ensembl | |
|
COSM3178819 rs2127846854 COSM4357440 |
905 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs2127846854 | 905 | G>V | No | Ensembl | |
| rs762475797 | 906 | T>A | No |
ExAC gnomAD |
|
| rs772890525 | 906 | T>I | No |
ExAC gnomAD |
|
| rs772890525 | 906 | T>N | No |
ExAC gnomAD |
|
| rs762475797 | 906 | T>P | No |
ExAC gnomAD |
|
| rs762475797 | 906 | T>S | No |
ExAC gnomAD |
|
| rs772890525 | 906 | T>S | No |
ExAC gnomAD |
|
| rs1562040318 | 907 | C>* | No |
TOPMed gnomAD |
|
| rs2127846837 | 907 | C>F | No | Ensembl | |
| rs2127846837 | 907 | C>S | No | Ensembl | |
| rs2127846839 | 907 | C>S | No | Ensembl | |
| rs2127846837 | 907 | C>Y | No | Ensembl | |
| rs1299934025 | 908 | S>C | No |
TOPMed gnomAD |
|
| rs1299934025 | 908 | S>G | No |
TOPMed gnomAD |
|
| rs1244617177 | 908 | S>N | No |
TOPMed gnomAD |
|
| rs769382910 | 908 | S>R | No |
ExAC gnomAD |
|
| rs1244617177 | 908 | S>T | No |
TOPMed gnomAD |
|
| rs1017335866 | 909 | R>G | No | Ensembl | |
| rs2127846815 | 909 | R>K | No | Ensembl | |
| rs2127846815 | 909 | R>T | No | Ensembl | |
| rs756054614 | 910 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs947671764 | 910 | P>S | No |
TOPMed gnomAD |
|
| rs947671764 | 910 | P>T | No |
TOPMed gnomAD |
|
| rs2127845640 | 911 | I>M | No | Ensembl | |
| rs1769902447 | 912 | S>G | No | Ensembl | |
| rs1198293940 | 915 | L>P | No | gnomAD | |
| rs1769902179 | 916 | D>G | No | TOPMed | |
| TCGA novel | 916 | D>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs149864613 | 917 | Q>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs1270494312 | 919 | T>S | No | gnomAD | |
| rs1769901642 | 920 | P>S | No | TOPMed | |
|
COSM3630589 COSM4895004 |
921 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749385662 | 922 | F>C | No |
ExAC gnomAD |
|
|
COSM3430959 COSM4946279 |
922 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 922 | F>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs139990635 | 923 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs752942156 | 924 | T>I | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 924 | T>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752942156 | 924 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs752942156 | 924 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs368531054 | 925 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs751918950 | 926 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs751918950 | 926 | C>S | No |
ExAC TOPMed gnomAD |
|
| rs1214794029 | 928 | V>A | No |
TOPMed gnomAD |
|
|
COSM4897253 COSM3630587 |
929 | G>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM4786995 COSM1446482 |
930 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM3876198 COSM3876197 |
930 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766896709 | 933 | Q>R | No |
ExAC gnomAD |
|
| rs763374008 | 934 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs890770968 | 935 | I>F | No |
TOPMed gnomAD |
|
| rs749890930 | 935 | I>N | No |
ExAC gnomAD |
|
| rs890770968 | 935 | I>V | No |
TOPMed gnomAD |
|
| rs1329501169 | 938 | E>Q | No | gnomAD | |
| rs764861058 | 939 | R>T | No |
ExAC gnomAD |
|
| rs2127845594 | 940 | Y>C | No | Ensembl | |
| rs1769899287 | 940 | Y>H | No | gnomAD | |
| rs1399866665 | 943 | N>S | No | gnomAD | |
| rs1216746396 | 944 | F>Y | No |
TOPMed gnomAD |
|
| rs151105732 | 945 | T>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1769898589 | 946 | A>T | No | TOPMed | |
| rs1279528179 | 947 | A>G | No | gnomAD | |
| rs776388575 | 948 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs374288532 | 950 | N>S | No |
ESP TOPMed gnomAD |
|
| rs141581954 | 951 | S>C | No |
TOPMed gnomAD |
|
|
COSM109723 COSM4484791 rs141581954 |
951 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
rs1243004793 COSM141172 |
952 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs745810696 | 952 | L>P | No |
ExAC gnomAD |
|
| rs1266503068 | 956 | A>V | No |
TOPMed gnomAD |
|
| rs1476264499 | 957 | R>K | No | TOPMed | |
| rs542454175 | 957 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2127845553 | 958 | M>I | No | Ensembl | |
| rs1351552549 | 959 | T>I | No |
TOPMed gnomAD |
|
| rs1351552549 | 959 | T>N | No |
TOPMed gnomAD |
|
| rs781371840 | 960 | I>T | No |
ExAC gnomAD |
|
| rs765208949 | 961 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs760252640 | 962 | D>V | No |
ExAC gnomAD |
|
| TCGA novel | 963 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1487534212 | 963 | V>M | No | Ensembl | |
| rs1769773936 | 964 | M>I | No | Ensembl | |
| rs1769774086 | 964 | M>K | No | Ensembl | |
| rs1286895663 | 964 | M>L | No | gnomAD | |
| rs1286895663 | 964 | M>V | No | gnomAD | |
| rs2127844081 | 966 | L>V | No | Ensembl | |
| rs267601169 | 967 | G>E | No | Ensembl | |
|
COSM4876784 COSM4876785 rs560174275 |
967 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA gnomAD |
| rs1397819533 | 968 | I>M | No | gnomAD | |
| TCGA novel | 969 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1422250111 | 972 | G>D | No |
TOPMed gnomAD |
|
| rs776759004 | 977 | I>M | No |
ExAC gnomAD |
|
| rs748213409 | 977 | I>T | No |
ExAC gnomAD |
|
|
COSM4786429 COSM1446480 |
978 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs374361032 | 978 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1186298360 | 978 | M>V | No |
TOPMed gnomAD |
|
| rs1244079200 | 981 | I>V | No | gnomAD | |
| rs1769771684 | 982 | Q>H | No | Ensembl | |
| rs1418929572 | 984 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs747218317 | 984 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs1219596340 | 984 | M>V | No | gnomAD | |
| rs1769770918 | 985 | R>K | No | TOPMed | |
| rs1292575307 | 987 | Q>R | No | gnomAD | |
| rs200425225 | 988 | M>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs758658280 | 990 | H>P | No |
ExAC TOPMed gnomAD |
|
| rs758658280 | 990 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs746280640 | 993 | G>* | No |
ExAC TOPMed gnomAD |
|
|
COSM274986 rs755519422 |
995 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs2127844016 | 997 | Q>* | No | Ensembl | |
| rs2127844016 | 997 | Q>E | No | Ensembl | |
| rs779546139 | 997 | Q>R | No |
ExAC gnomAD |
|
| TCGA novel | 999 | V>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q15375
14 regional properties for Q15375
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 633 - 894 | IPR000719 |
| domain | Ephrin receptor ligand binding domain | 32 - 210 | IPR001090 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 633 - 888 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 186 - 206 | IPR001426-1 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 250 - 270 | IPR001426-2 |
| domain | Sterile alpha motif domain | 920 - 987 | IPR001660 |
| domain | Fibronectin type III | 331 - 441 | IPR003961-1 |
| domain | Fibronectin type III | 442 - 537 | IPR003961-2 |
| active_site | Tyrosine-protein kinase, active site | 754 - 766 | IPR008266 |
| domain | Tyrosine-protein kinase ephrin type A/B receptor-like | 271 - 308 | IPR011641 |
| binding_site | Protein kinase, ATP binding site | 639 - 665 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 633 - 890 | IPR020635 |
| domain | Ephrin receptor, transmembrane domain | 560 - 630 | IPR027936 |
| domain | Ephrin type-A receptor 7, ligand binding domain | 30 - 206 | IPR034283 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.1 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| dendrite | A neuron projection that has a short, tapering, morphology. Dendrites receive and integrate signals from other neurons or from sensory stimuli, and conduct nerve impulses towards the axon or the cell body. In most neurons, the impulse is conveyed from dendrites to axon via the cell body, but in some types of unipolar neuron, the impulse does not travel via the cell body. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| axon guidance receptor activity | Combining with an extracellular messenger and transmitting the signal from one side of the membrane to the other to results in a change in cellular activity involved in axon guidance. |
| chemorepellent activity | Providing the environmental signal that initiates the directed movement of a motile cell or organism towards a lower concentration of that signal. |
| GPI-linked ephrin receptor activity | Combining with a GPI-anchored ephrin to initiate a change in cell activity. |
| growth factor binding | Binding to a growth factor, proteins or polypeptides that stimulate a cell or organism to grow or proliferate. |
| protein tyrosine kinase activity | Catalysis of the reaction |
| transmembrane-ephrin receptor activity | Combining with a transmembrane ephrin to initiate a change in cell activity. |
20 GO annotations of biological process
| Name | Definition |
|---|---|
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| branching morphogenesis of a nerve | The process in which the anatomical structures of branches in a nerve are generated and organized. This term refers to an anatomical structure (nerve) not a cell (neuron). |
| ephrin receptor signaling pathway | The series of molecular signals initiated by ephrin binding to its receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| modulation of chemical synaptic transmission | Any process that modulates the frequency or amplitude of synaptic transmission, the process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse. Amplitude, in this case, refers to the change in postsynaptic membrane potential due to a single instance of synaptic transmission. |
| negative chemotaxis | The directed movement of a motile cell or organism towards a lower concentration of a chemical. |
| negative regulation of collateral sprouting | Any process that stops, prevents, or reduces the frequency, rate or extent of collateral sprouting. |
| negative regulation of synapse assembly | Any process that stops, prevents, or reduces the frequency, rate or extent of synapse assembly, the aggregation, arrangement and bonding together of a set of components to form a synapse. |
| nephric duct morphogenesis | The process in which the anatomical structures of the nephric duct are generated and organized. A nephric duct is a tube that drains a primitive kidney. |
| neuron apoptotic process | Any apoptotic process in a neuron, the basic cellular unit of nervous tissue. Each neuron consists of a body, an axon, and dendrites. Their purpose is to receive, conduct, and transmit impulses in the nervous system. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of neuron apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death of neurons by apoptotic process. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| regulation of cell-cell adhesion | Any process that modulates the frequency, rate or extent of attachment of a cell to another cell. |
| regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that modulates the activity of a cysteine-type endopeptidase involved in apoptosis. |
| regulation of ERK1 and ERK2 cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| regulation of peptidyl-tyrosine phosphorylation | Any process that modulates the frequency, rate or extent of the phosphorylation of peptidyl-tyrosine. |
| regulation of postsynapse organization | Any process that modulates the physical form of a postsynapse. |
| regulation of protein autophosphorylation | Any process that modulates the frequency, rate or extent of addition of the phosphorylation by a protein of one or more of its own residues. |
| retinal ganglion cell axon guidance | The process in which the migration of an axon growth cone of a retinal ganglion cell (RGC) is directed to its target in the brain in response to a combination of attractive and repulsive cues. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P21709 | EPHA1 | Ephrin type-A receptor 1 | Homo sapiens (Human) | SS |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| P54756 | EPHA5 | Ephrin type-A receptor 5 | Homo sapiens (Human) | SS |
| Q9UF33 | EPHA6 | Ephrin type-A receptor 6 | Homo sapiens (Human) | SS |
| P54762 | EPHB1 | Ephrin type-B receptor 1 | Homo sapiens (Human) | SS |
| P29322 | EPHA8 | Ephrin type-A receptor 8 | Homo sapiens (Human) | SS |
| P29320 | EPHA3 | Ephrin type-A receptor 3 | Homo sapiens (Human) | PR |
| P54764 | EPHA4 | Ephrin type-A receptor 4 | Homo sapiens (Human) | SS |
| P54753 | EPHB3 | Ephrin type-B receptor 3 | Homo sapiens (Human) | SS |
| Q5JZY3 | EPHA10 | Ephrin type-A receptor 10 | Homo sapiens (Human) | SS |
| O15197 | EPHB6 | Ephrin type-B receptor 6 | Homo sapiens (Human) | SS |
| P29323 | EPHB2 | Ephrin type-B receptor 2 | Homo sapiens (Human) | EV |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| O08644 | Ephb6 | Ephrin type-B receptor 6 | Mus musculus (Mouse) | PR |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| O61460 | vab-1 | Ephrin receptor 1 | Caenorhabditis elegans | SS |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E991 | PRK6 | Pollen receptor-like kinase 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FXF2 | RKF1 | Probable LRR receptor-like serine/threonine-protein kinase RFK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O13147 | ephb3 | Ephrin type-B receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73878 | ephb4b | Ephrin type-B receptor 4b | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVFQTRYPSW | IILCYIWLLR | FAHTGEAQAA | KEVLLLDSKA | QQTELEWISS | PPNGWEEISG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LDENYTPIRT | YQVCQVMEPN | QNNWLRTNWI | SKGNAQRIFV | ELKFTLRDCN | SLPGVLGTCK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ETFNLYYYET | DYDTGRNIRE | NLYVKIDTIA | ADESFTQGDL | GERKMKLNTE | VREIGPLSKK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GFYLAFQDVG | ACIALVSVKV | YYKKCWSIIE | NLAIFPDTVT | GSEFSSLVEV | RGTCVSSAEE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EAENAPRMHC | SAEGEWLVPI | GKCICKAGYQ | QKGDTCEPCG | RGFYKSSSQD | LQCSRCPTHS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| FSDKEGSSRC | ECEDGYYRAP | SDPPYVACTR | PPSAPQNLIF | NINQTTVSLE | WSPPADNGGR |
| 370 | 380 | 390 | 400 | 410 | 420 |
| NDVTYRILCK | RCSWEQGECV | PCGSNIGYMP | QQTGLEDNYV | TVMDLLAHAN | YTFEVEAVNG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VSDLSRSQRL | FAAVSITTGQ | AAPSQVSGVM | KERVLQRSVE | LSWQEPEHPN | GVITEYEIKY |
| 490 | 500 | 510 | 520 | 530 | 540 |
| YEKDQRERTY | STVKTKSTSA | SINNLKPGTV | YVFQIRAFTA | AGYGNYSPRL | DVATLEEATG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| KMFEATAVSS | EQNPVIIIAV | VAVAGTIILV | FMVFGFIIGR | RHCGYSKADQ | EGDEELYFHF |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KFPGTKTYID | PETYEDPNRA | VHQFAKELDA | SCIKIERVIG | AGEFGEVCSG | RLKLPGKRDV |
| 670 | 680 | 690 | 700 | 710 | 720 |
| AVAIKTLKVG | YTEKQRRDFL | CEASIMGQFD | HPNVVHLEGV | VTRGKPVMIV | IEFMENGALD |
| 730 | 740 | 750 | 760 | 770 | 780 |
| AFLRKHDGQF | TVIQLVGMLR | GIAAGMRYLA | DMGYVHRDLA | ARNILVNSNL | VCKVSDFGLS |
| 790 | 800 | 810 | 820 | 830 | 840 |
| RVIEDDPEAV | YTTTGGKIPV | RWTAPEAIQY | RKFTSASDVW | SYGIVMWEVM | SYGERPYWDM |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SNQDVIKAIE | EGYRLPAPMD | CPAGLHQLML | DCWQKERAER | PKFEQIVGIL | DKMIRNPNSL |
| 910 | 920 | 930 | 940 | 950 | 960 |
| KTPLGTCSRP | ISPLLDQNTP | DFTTFCSVGE | WLQAIKMERY | KDNFTAAGYN | SLESVARMTI |
| 970 | 980 | 990 | |||
| EDVMSLGITL | VGHQKKIMSS | IQTMRAQMLH | LHGTGIQV |