Q15306
Gene name |
IRF4 (MUM1) |
Protein name |
Interferon regulatory factor 4 |
Names |
IRF-4, Lymphocyte-specific interferon regulatory factor, LSIRF, Multiple myeloma oncogene 1, NF-EM5 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:3662 |
EC number |
|
Protein Class |
|
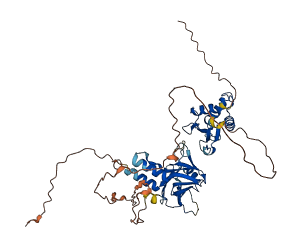
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
249-418 (IFN association domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
References
- Remesh SG et al. (2015) "Structural Studies of IRF4 Reveal a Flexible Autoinhibitory Region and a Compact Linker Domain", The Journal of biological chemistry, 290, 27779-90
- Brass AL et al. (1999) "Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers", The EMBO journal, 18, 977-91
- Brass AL et al. (1996) "Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1", Genes & development, 10, 2335-47
- Sathish N et al. (2011) "Mechanisms of autoinhibition of IRF-7 and a probable model for inactivation of IRF-7 by Kaposi's sarcoma-associated herpesvirus protein ORF45", The Journal of biological chemistry, 286, 746-56
- Ning S et al. (2011) "IRF7: activation, regulation, modification and function", Genes and immunity, 12, 399-414
- Lin R et al. (1999) "Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains", Molecular and cellular biology, 19, 2465-74
- Shah M et al. (2015) "In silico mechanistic analysis of IRF3 inactivation and high-risk HPV E6 species-dependent drug response", Scientific reports, 5, 13446
- Liu S et al. (2015) "Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation", Science (New York, N.Y.), 347, aaa2630
- Barnes BJ et al. (2002) "Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes", Molecular and cellular biology, 22, 5721-40
Autoinhibited structure

Activated structure

8 structures for Q15306
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DLL | NMR | - | A | 23-130 | PDB |
| 6TD4 | X-ray | 171 A | A | 20-132 | PDB |
| 7JM4 | X-ray | 295 A | A/B/G/H | 21-129 | PDB |
| 7O56 | X-ray | 260 A | A/B/C | 20-139 | PDB |
| 7OGS | X-ray | 237 A | A/B/E/F | 20-139 | PDB |
| 7OOT | X-ray | 225 A | A/B | 20-139 | PDB |
| 7RH2 | X-ray | 247 A | A/B/G/H | 21-129 | PDB |
| AF-Q15306-F1 | Predicted | AlphaFoldDB |
235 variants for Q15306
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000714777 CA133337128 rs901736609 |
363 | Q>H | Skin/hair/eye pigmentation, variation in, 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
COSM220747 rs891224445 CA133328978 |
2 | N>S | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs1452883883 CA362546230 |
4 | E>* | No |
ClinGen gnomAD |
|
|
CA362546232 rs1581220805 |
4 | E>G | No |
ClinGen Ensembl |
|
|
CA133329000 rs564941413 |
5 | G>D | No |
ClinGen Ensembl |
|
|
rs1296646377 CA362546242 |
6 | G>R | No |
ClinGen gnomAD |
|
|
rs769695946 CA3612604 |
7 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362546252 rs1251757585 |
8 | R>G | No |
ClinGen gnomAD |
|
|
rs139884486 CA3612606 |
8 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3612605 rs139884486 |
8 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1581220840 CA362546259 |
9 | G>D | No |
ClinGen Ensembl |
|
|
rs1285362149 CA362546255 |
9 | G>S | No |
ClinGen TOPMed |
|
|
rs1403333371 CA362546261 |
10 | G>R | No |
ClinGen TOPMed |
|
|
rs546956488 CA133329015 |
12 | F>L | No |
ClinGen 1000Genomes TOPMed |
|
|
CA362546293 rs1265068205 |
14 | M>R | No |
ClinGen gnomAD |
|
|
rs1343907006 CA362546318 |
18 | S>G | No |
ClinGen TOPMed |
|
|
rs1000698747 CA362546321 |
18 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1000698747 CA133329066 COSM1731950 |
18 | S>N | NS [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
COSM220626 CA362546320 rs1000698747 |
18 | S>T | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs538907751 CA3612611 |
35 | G>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1581220917 CA362546448 |
36 | K>T | No |
ClinGen Ensembl |
|
|
CA3612613 rs755738102 |
38 | P>L | No |
ClinGen ExAC |
|
|
CA3612612 rs369688140 |
38 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1230948269 CA362546468 |
39 | G>E | No |
ClinGen gnomAD |
|
|
CA3612615 rs751037944 |
41 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA362546489 rs1208372867 |
42 | W>* | No |
ClinGen gnomAD |
|
|
CA133329147 rs934099606 |
45 | E>Q | No |
ClinGen TOPMed |
|
|
rs780297970 CA3612618 |
46 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1431984073 CA362546524 |
47 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs781441118 CA3612621 |
53 | P>H | No |
ClinGen ExAC |
|
|
CA133329252 rs868350590 |
66 | E>G | No |
ClinGen Ensembl |
|
|
rs1286533547 CA362546707 |
72 | K>R | No |
ClinGen gnomAD |
|
|
CA362546788 rs1302327519 |
82 | R>* | No |
ClinGen gnomAD |
|
|
rs1377317453 CA362546789 |
82 | R>Q | No |
ClinGen gnomAD |
|
|
rs1454761481 CA362546812 |
85 | I>M | No |
ClinGen gnomAD |
|
|
rs144593192 CA3612651 |
88 | P>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs202124383 CA362546831 |
88 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3612652 rs202124383 |
88 | P>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA133330899 rs144593192 |
88 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA362546838 rs1226872247 |
89 | D>E | No |
ClinGen gnomAD |
|
|
CA362546866 rs1303460354 |
93 | W>C | No |
ClinGen TOPMed |
|
|
CA362546881 COSM1173222 rs1459743512 |
96 | R>C | oesophagus [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA3612657 COSM1754738 rs767879003 |
100 | A>T | urinary_tract [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs773571916 CA133330921 |
105 | N>D | No |
ClinGen Ensembl |
|
|
rs200303421 CA3612658 |
105 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612660 rs374588785 |
110 | L>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs944381620 CA133330939 |
113 | R>Q | No |
ClinGen Ensembl |
|
|
rs753842732 COSM1162263 CA3612661 |
113 | R>W | pancreas [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1320369594 CA362547049 |
120 | D>E | No |
ClinGen TOPMed |
|
|
rs745876631 CA3612664 |
126 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1272287601 CA362547103 |
128 | V>D | No |
ClinGen gnomAD |
|
|
rs771936972 CA3612665 |
128 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA362547139 rs1320732051 |
134 | K>E | No |
ClinGen gnomAD |
|
|
CA3612686 rs758443557 |
136 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA3612687 rs780187356 |
137 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA362547177 rs1490819568 |
138 | Q>* | No |
ClinGen gnomAD |
|
|
CA3612689 rs754886586 |
139 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA3612691 rs11557493 |
141 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs769115445 CA3612692 |
144 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA3612694 rs115613112 |
146 | M>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA362547245 rs1300078693 |
148 | M>T | No |
ClinGen gnomAD |
|
|
rs1406059165 CA362547242 |
148 | M>V | No |
ClinGen gnomAD |
|
|
CA3612695 rs73717071 |
149 | S>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA133331838 rs201185528 |
150 | H>Y | No |
ClinGen 1000Genomes |
|
|
rs1351148355 CA362547266 |
151 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1289314958 CA362547274 |
152 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1245046920 CA362547271 |
152 | Y>H | No |
ClinGen gnomAD |
|
|
CA362547284 rs764577689 |
154 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764577689 CA3612698 |
154 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1368848977 CA362547294 |
155 | T>K | No |
ClinGen TOPMed |
|
|
CA3612699 rs776661032 |
155 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs755460615 CA3612700 |
156 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612702 rs750505973 |
158 | Y>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1561712197 CA362547318 |
159 | P>H | No |
ClinGen Ensembl |
|
|
CA362547324 rs1472591082 |
160 | S>L | No |
ClinGen gnomAD |
|
|
CA3612705 rs766413794 |
162 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs1236751116 CA362547374 |
166 | V>I | No |
ClinGen TOPMed |
|
|
rs767166788 CA3612726 |
167 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs200611169 CA133332948 |
167 | H>R | No |
ClinGen 1000Genomes TOPMed |
|
|
rs1301653439 CA362547393 |
169 | Y>H | No |
ClinGen TOPMed |
|
|
CA362547406 rs1324660759 |
170 | M>I | No |
ClinGen gnomAD |
|
|
CA362547414 rs1270868534 |
171 | M>I | No |
ClinGen gnomAD |
|
|
CA3612728 rs755905242 |
171 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs1236233284 CA362547410 |
171 | M>V | No |
ClinGen TOPMed |
|
|
rs777399667 CA3612729 |
172 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1322864517 CA362547419 |
172 | P>S | No |
ClinGen gnomAD |
|
|
rs1581224450 CA362547433 |
174 | L>R | No |
ClinGen Ensembl |
|
|
rs1461930692 CA362547434 COSM3830471 |
175 | D>N | breast [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA3612731 rs756699452 |
176 | R>* | No |
ClinGen ExAC |
|
|
rs999945656 CA133332970 COSM1179728 |
176 | R>Q | prostate [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1260048530 CA362547475 |
180 | D>E | No |
ClinGen gnomAD |
|
|
CA3612732 rs778403209 |
180 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA3612733 rs747437180 |
181 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs866516667 CA133332974 |
181 | Y>H | No |
ClinGen TOPMed |
|
|
COSM2153954 rs781586995 CA3612735 |
182 | V>I | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA3612737 rs770110760 COSM1546336 |
185 | Q>H | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1431065750 CA362547513 |
186 | P>L | No |
ClinGen gnomAD |
|
|
CA362547518 rs1322810804 |
187 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
CA362547527 rs1469969945 |
188 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs771094772 CA3612740 |
190 | I>N | No |
ClinGen ExAC gnomAD |
|
|
rs763272146 CA3612739 |
190 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA3612741 rs774593906 |
191 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA362547563 rs1393717852 |
194 | C>R | No |
ClinGen TOPMed |
|
|
rs1285265620 CA362547577 |
196 | M>V | No |
ClinGen TOPMed gnomAD |
|
|
CA3612744 rs372869810 |
197 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs372869810 CA362547589 |
197 | T>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3612746 rs763891187 |
198 | F>S | No |
ClinGen ExAC gnomAD |
|
|
rs1487028591 CA362547605 |
200 | P>S | No |
ClinGen gnomAD |
|
|
rs1206239895 CA362547611 |
201 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA3612747 rs753427028 COSM1079040 |
201 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs753427028 CA362547613 |
201 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM321045 rs143144957 CA3612749 |
202 | G>S | lung large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs1270389563 CA362547668 |
207 | G>S | No |
ClinGen gnomAD |
|
|
CA3612751 rs757910134 |
208 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs893823313 CA133334351 |
213 | G>V | No |
ClinGen TOPMed |
|
|
rs538810711 CA3612778 |
214 | C>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1293066909 CA362547917 |
216 | V>M | No |
ClinGen gnomAD |
|
|
rs746092245 CA3612779 |
217 | T>R | No |
ClinGen ExAC gnomAD |
|
|
CA362547989 rs1561714266 |
224 | A>T | No |
ClinGen Ensembl |
|
|
CA362547996 rs1321465734 |
225 | P>T | No |
ClinGen gnomAD |
|
|
CA3612780 rs147454166 |
230 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA133334373 rs62389395 |
230 | A>V | No |
ClinGen Ensembl |
|
|
rs375133421 CA3612782 |
232 | G>R | No |
ClinGen 1000Genomes ESP ExAC gnomAD |
|
|
rs761579830 CA3612785 |
233 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs776593233 CA3612784 |
233 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1561714310 CA362548102 |
234 | P>H | No |
ClinGen Ensembl |
|
|
CA3612786 rs765027259 |
235 | T>K | No |
ClinGen ExAC gnomAD |
|
|
rs149628278 CA133334433 |
239 | I>V | No |
ClinGen ESP |
|
|
rs772851591 CA362548185 |
242 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs772851591 CA3612787 |
242 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA133334442 rs991648246 |
242 | A>V | No |
ClinGen gnomAD |
|
|
CA362548194 COSM1079069 rs1356308443 |
243 | E>K | endometrium central_nervous_system [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA3612789 rs765864636 |
245 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA362548240 rs751010198 |
246 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1079070 CA3612790 rs751010198 |
246 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs758839744 CA3612814 |
249 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs199834880 CA133336799 |
251 | R>Q | No |
ClinGen gnomAD |
|
|
rs1198842534 CA362548616 |
254 | I>V | No |
ClinGen gnomAD |
|
|
rs148760922 CA3612816 |
256 | L>M | No |
ClinGen ESP ExAC gnomAD |
|
|
CA133336836 rs975308028 |
257 | Y>F | No |
ClinGen TOPMed gnomAD |
|
|
rs781276700 CA3612818 |
259 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA362548679 rs1407719860 |
259 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs752393433 CA3612824 |
272 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA3612827 rs139858073 |
275 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs771669183 CA3612826 COSM170244 |
275 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA3612829 rs765634575 |
278 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA362548822 rs1446497140 |
279 | G>R | No |
ClinGen TOPMed |
|
|
rs751008894 CA3612830 |
281 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612833 rs114916515 |
283 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs767774669 CA362548857 |
284 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767774669 CA3612835 |
284 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362548858 rs1323096518 |
284 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1179357950 CA362548889 |
288 | D>E | No |
ClinGen gnomAD |
|
|
CA362548896 rs1222738803 |
289 | Q>H | No |
ClinGen gnomAD |
|
|
rs1224284590 CA362548928 |
294 | Y>C | No |
ClinGen TOPMed |
|
|
rs1356593229 CA362548937 |
296 | E>K | No |
ClinGen TOPMed |
|
|
CA3612836 rs140099868 |
298 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1050972 CA133336939 |
300 | Q>H | No |
ClinGen Ensembl |
|
|
rs756281792 CA3612837 |
304 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA362549033 rs1302019454 |
309 | S>R | No |
ClinGen TOPMed |
|
|
CA133336967 rs780902340 |
315 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612840 rs780902340 |
315 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612842 rs745470687 |
317 | L>I | No |
ClinGen ExAC gnomAD |
|
|
CA133337034 rs775326241 |
320 | A>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3612844 rs775326241 |
320 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1210576093 CA362549113 |
322 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA3612847 rs773521348 |
325 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA3612848 rs201105575 |
326 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA133337073 rs1050974 |
333 | R>T | No |
ClinGen Ensembl |
|
|
CA362549241 rs1182121626 |
338 | G>R | No |
ClinGen gnomAD |
|
|
rs1581227987 CA362549251 |
339 | P>T | No |
ClinGen Ensembl |
|
|
CA3612852 rs200311468 |
341 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA133337105 rs888989341 |
342 | L>P | No |
ClinGen TOPMed |
|
|
rs756088477 CA3612854 |
344 | N>H | No |
ClinGen ExAC gnomAD |
|
|
rs753680565 CA3612856 |
345 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362549332 rs1400777715 |
346 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA3612857 COSM1079083 rs199596593 |
346 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
rs1400777715 CA362549334 |
346 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA362549353 rs1334437649 |
348 | N>H | No |
ClinGen gnomAD |
|
|
CA362549359 rs1561716191 |
348 | N>T | No |
ClinGen Ensembl |
|
|
rs199919569 CA3612859 |
353 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362549454 rs1343858008 |
355 | T>A | No |
ClinGen TOPMed |
|
|
CA3612860 rs555752097 |
357 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA3612861 rs779841481 |
362 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1380305124 CA362549560 |
366 | S>L | No |
ClinGen gnomAD |
|
|
rs568315642 CA362550136 |
370 | A>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1049908319 CA133339845 |
370 | A>T | No |
ClinGen TOPMed |
|
|
rs568315642 CA3612886 |
370 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1581230749 CA362550203 |
373 | H>P | No |
ClinGen Ensembl |
|
|
rs775948276 CA3612888 |
373 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1169247876 CA362550246 |
375 | G>S | No |
ClinGen gnomAD |
|
|
CA3612890 rs768893830 |
376 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs377483798 CA3612891 |
376 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs377483798 CA362550262 |
376 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs377483798 CA3612892 |
376 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA362550291 rs1421475486 |
378 | L>Q | No |
ClinGen gnomAD |
|
|
rs1159107414 CA362550305 |
379 | P>T | No |
ClinGen gnomAD |
|
|
CA3612894 rs750322004 |
382 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA362550348 rs1413303208 |
382 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs142143231 CA133339878 |
384 | T>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs142143231 CA3612895 |
384 | T>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3612897 rs751335149 |
390 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201428294 CA3612898 |
394 | P>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs201428294 CA3612899 |
394 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA362550703 rs141640145 |
404 | H>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1357924165 CA362550696 |
404 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs199763142 CA3612935 |
411 | R>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1301548815 CA362550845 |
412 | Q>L | No |
ClinGen TOPMed |
|
|
rs1400229165 CA362550860 |
414 | Y>H | No |
ClinGen gnomAD |
|
|
CA362550969 rs1220781475 |
422 | G>R | No |
ClinGen gnomAD |
|
|
CA133342220 rs867625247 |
423 | H>N | No |
ClinGen Ensembl |
|
|
CA3612938 rs760504688 |
424 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763733876 CA3612939 |
425 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1214308590 CA362551027 |
427 | G>A | No |
ClinGen gnomAD |
|
|
CA362551025 rs1581232368 |
427 | G>C | No |
ClinGen Ensembl |
|
|
CA133342226 rs934318315 |
428 | Y>C | No |
ClinGen Ensembl |
|
|
rs1227685527 CA362551039 |
429 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA133342229 rs908661692 |
429 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs756736434 CA3612941 |
434 | I>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA133342244 rs892912014 |
434 | I>M | No |
ClinGen Ensembl |
|
|
CA362551074 rs756736434 |
434 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA133342259 rs1011269643 |
437 | P>L | No |
ClinGen TOPMed |
|
|
rs1488875183 CA362551133 |
439 | D>G | No |
ClinGen TOPMed |
|
|
rs1160900661 CA362551144 |
440 | Y>F | No |
ClinGen gnomAD |
|
|
CA362551138 rs1438413858 |
440 | Y>N | No |
ClinGen gnomAD |
|
|
rs752069005 CA3612944 |
441 | H>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA133342303 rs752069005 |
441 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1322275605 CA362551176 |
443 | S>C | No |
ClinGen gnomAD |
|
|
CA3612946 rs559734928 |
444 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs748516736 CA3612947 |
445 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA3612948 rs756602021 |
445 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756602021 CA3612949 |
445 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362551192 rs1237888981 |
446 | H>Y | No |
ClinGen gnomAD |
|
|
CA3612950 rs749474191 |
447 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA362551231 rs1354319999 |
448 | S>F | No |
ClinGen gnomAD |
1 associated diseases with Q15306
[MIM: 254500]: Multiple myeloma (MM)
A malignant tumor of plasma cells usually arising in the bone marrow and characterized by diffuse involvement of the skeletal system, hyperglobulinemia, Bence-Jones proteinuria and anemia. Complications of multiple myeloma are bone pain, hypercalcemia, renal failure and spinal cord compression. The aberrant antibodies that are produced lead to impaired humoral immunity and patients have a high prevalence of infection. Amyloidosis may develop in some patients. Multiple myeloma is part of a spectrum of diseases ranging from monoclonal gammopathy of unknown significance (MGUS) to plasma cell leukemia. {ECO:0000269|PubMed:9326949}. Note=The gene represented in this entry may be involved in disease pathogenesis. A chromosomal aberration involving IRF4 has been found in multiple myeloma. Translocation t(6;14)(p25;q32) with the IgH locus.
Without disease ID
- A malignant tumor of plasma cells usually arising in the bone marrow and characterized by diffuse involvement of the skeletal system, hyperglobulinemia, Bence-Jones proteinuria and anemia. Complications of multiple myeloma are bone pain, hypercalcemia, renal failure and spinal cord compression. The aberrant antibodies that are produced lead to impaired humoral immunity and patients have a high prevalence of infection. Amyloidosis may develop in some patients. Multiple myeloma is part of a spectrum of diseases ranging from monoclonal gammopathy of unknown significance (MGUS) to plasma cell leukemia. {ECO:0000269|PubMed:9326949}. Note=The gene represented in this entry may be involved in disease pathogenesis. A chromosomal aberration involving IRF4 has been found in multiple myeloma. Translocation t(6;14)(p25;q32) with the IgH locus.
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleosome | A complex comprised of DNA wound around a multisubunit core and associated proteins, which forms the primary packing unit of DNA into higher order structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| transcription coactivator activity | A transcription coregulator activity that activates or increases the transcription of specific gene sets via binding to a DNA-bound DNA-binding transcription factor, either on its own or as part of a complex. Coactivators often act by altering chromatin structure and modifications. For example, one class of transcription coactivators modifies chromatin structure through covalent modification of histones. A second class remodels the conformation of chromatin in an ATP-dependent fashion. A third class modulates interactions of DNA-bound DNA-binding transcription factors with other transcription coregulators. A fourth class of coactivator activity is the bridging of a DNA-binding transcription factor to the general (basal) transcription machinery. The Mediator complex, which bridges sequence-specific DNA binding transcription factors and RNA polymerase, is also a transcription coactivator. |
18 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response to protozoan | Reactions triggered in response to the presence of a protozoan that act to protect the cell or organism. |
| histone H3 acetylation | The modification of histone H3 by the addition of an acetyl group. |
| histone H4 acetylation | The modification of histone H4 by the addition of an acetyl group. |
| immune system process | Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats. |
| myeloid dendritic cell differentiation | The process in which a monocyte acquires the specialized features of a dendritic cell, an immunocompetent cell of the lymphoid and hemopoietic systems and skin. |
| negative regulation of toll-like receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate, or extent of toll-like receptor signaling pathway. |
| positive regulation of cold-induced thermogenesis | Any process that activates or increases the frequency, rate or extent of cold-induced thermogenesis. |
| positive regulation of DNA binding | Any process that increases the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of interleukin-10 production | Any process that activates or increases the frequency, rate, or extent of interleukin-10 production. |
| positive regulation of interleukin-13 production | Any process that activates or increases the frequency, rate, or extent of interleukin-13 production. |
| positive regulation of interleukin-2 production | Any process that activates or increases the frequency, rate, or extent of interleukin-2 production. |
| positive regulation of interleukin-4 production | Any process that activates or increases the frequency, rate, or extent of interleukin-4 production. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| regulation of T-helper cell differentiation | Any process that modulates the frequency, rate or extent of T-helper cell differentiation. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
| T-helper 17 cell lineage commitment | The process in which a CD4-positive, alpha-beta T cell becomes committed to becoming a T-helper 17 cell, a CD4-positive, alpha-beta T cell with the phenotype RORgamma-t-positive that produces IL-17. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q00978 | IRF9 | Interferon regulatory factor 9 | Homo sapiens (Human) | PR |
| Q13568 | IRF5 | Interferon regulatory factor 5 | Homo sapiens (Human) | EV |
| Q14653 | IRF3 | Interferon regulatory factor 3 | Homo sapiens (Human) | EV |
| Q92985 | IRF7 | Interferon regulatory factor 7 | Homo sapiens (Human) | EV |
| Q02556 | IRF8 | Interferon regulatory factor 8 | Homo sapiens (Human) | PR |
| Q64287 | Irf4 | Interferon regulatory factor 4 | Mus musculus (Mouse) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNLEGGGRGG | EFGMSAVSCG | NGKLRQWLID | QIDSGKYPGL | VWENEEKSIF | RIPWKHAGKQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DYNREEDAAL | FKAWALFKGK | FREGIDKPDP | PTWKTRLRCA | LNKSNDFEEL | VERSQLDISD |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PYKVYRIVPE | GAKKGAKQLT | LEDPQMSMSH | PYTMTTPYPS | LPAQQVHNYM | MPPLDRSWRD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YVPDQPHPEI | PYQCPMTFGP | RGHHWQGPAC | ENGCQVTGTF | YACAPPESQA | PGVPTEPSIR |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SAEALAFSDC | RLHICLYYRE | ILVKELTTSS | PEGCRISHGH | TYDASNLDQV | LFPYPEDNGQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RKNIEKLLSH | LERGVVLWMA | PDGLYAKRLC | QSRIYWDGPL | ALCNDRPNKL | ERDQTCKLFD |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TQQFLSELQA | FAHHGRSLPR | FQVTLCFGEE | FPDPQRQRKL | ITAHVEPLLA | RQLYYFAQQN |
| 430 | 440 | 450 | |||
| SGHFLRGYDL | PEHISNPEDY | HRSIRHSSIQ | E |