Q14318
Gene name |
FKBP8 (FKBP38) |
Protein name |
Peptidyl-prolyl cis-trans isomerase FKBP8 |
Names |
PPIase FKBP8, 38 kDa FK506-binding protein, 38 kDa FKBP, FKBP-38, hFKBP38, FK506-binding protein 8, FKBP-8, FKBPR38, Rotamase |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23770 |
EC number |
5.2.1.8: Cis-trans isomerases |
Protein Class |
PEPTIDYLPROLYL ISOMERASE (PTHR46512) |
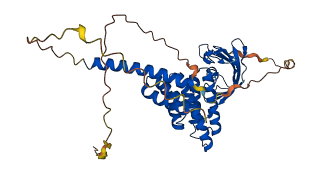
Descriptions
FK506 binding protein 38 (FKBP38) is a multi-domain protein containing a peptidyl-prolyl cis-trans isomerase domain (PPlase) domain followed by tetratricopeptide repeats (TPR), and transmembrane domain. FKBP38 regulates apoptosis through unique interactions with multiple regulators including Bcl-2. The N-terminal extension preceding the catalytic domain of FKBP38 has an autoinhibitory activity. The core isomerase activity of FKBP38 is inhibited by transient interactions involving the flexible N-terminal extension that precedes the catalytic domain. CaM/Ca2+ binds to the N-terminal extension and thereby releases the autoinhibitory contacts between the N-terminal extension and the catalytic domain.
Autoinhibitory domains (AIDs)
Target domain |
115-204 (Peptidyl-prolyl cis-trans isomerase domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure
Activated structure

253 variants for Q14318
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000850453 CA404825348 rs1485300085 |
241 | A>T | Marfanoid habitus and intellectual disability [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs543806511 CA9313387 |
3 | S>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA404830909 rs1243784318 |
7 | P>R | No |
ClinGen gnomAD |
|
|
CA404830865 rs1316686457 |
10 | P>S | No |
ClinGen gnomAD |
|
|
rs1444051981 CA404830771 |
15 | P>R | No |
ClinGen gnomAD |
|
|
rs11541428 CA306215528 COSM993123 |
16 | A>T | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs371498399 CA9313381 |
17 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs376029668 CA9313380 |
20 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1437565472 CA404830677 |
21 | L>P | No |
ClinGen gnomAD |
|
|
CA9313376 rs375921391 |
22 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1404448556 CA404830649 |
23 | D>N | No |
ClinGen gnomAD |
|
|
CA404830602 rs933275119 |
24 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
CA404830593 rs1481834096 |
25 | E>K | No |
ClinGen TOPMed |
|
|
rs1481834096 CA404830600 |
25 | E>Q | No |
ClinGen TOPMed |
|
|
rs1438019537 CA404830494 |
28 | D>E | No |
ClinGen gnomAD |
|
|
CA9313372 rs763260727 |
32 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763260727 CA404830432 |
32 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1489317297 CA404830386 |
33 | A>G | No |
ClinGen gnomAD |
|
|
CA404830384 rs1489317297 |
33 | A>V | No |
ClinGen gnomAD |
|
|
CA306215496 rs933677129 |
34 | E>K | No |
ClinGen TOPMed |
|
|
CA404829963 rs933677129 |
34 | E>Q | No |
ClinGen TOPMed |
|
|
CA9313369 rs760028266 |
34 | E>V | No |
ClinGen ExAC gnomAD |
|
|
rs776172610 COSM1564615 CA9313368 |
35 | G>D | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
rs1272735468 CA404829933 |
36 | E>A | No |
ClinGen Ensembl |
|
|
CA9313364 rs772769071 |
41 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771709504 CA9313362 |
44 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404829688 rs1233902961 |
47 | D>G | No |
ClinGen Ensembl |
|
|
CA404829681 rs1600540423 |
48 | D>A | No |
ClinGen Ensembl |
|
|
rs140276721 CA9313358 |
48 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1333540563 CA404829621 |
51 | E>G | No |
ClinGen TOPMed |
|
|
CA9313357 rs755032883 |
53 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA404829571 rs1176669010 |
54 | P>A | No |
ClinGen gnomAD |
|
|
rs749228796 CA9313356 |
54 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM389279 rs1484076170 CA404829489 |
58 | M>I | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1190477085 CA404829496 |
58 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1238967082 CA404829455 |
60 | Q>E | No |
ClinGen gnomAD |
|
|
CA404829434 rs751621562 |
60 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
CA9313352 rs764269613 |
61 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA9313350 rs537148195 |
62 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs759836176 CA9313348 |
63 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404829232 rs1310946899 |
69 | P>R | No |
ClinGen gnomAD |
|
|
CA306215433 rs1043736516 |
70 | G>W | No |
ClinGen Ensembl |
|
|
rs371852654 CA9313344 |
74 | R>G | No |
ClinGen ESP ExAC gnomAD |
|
|
rs771664117 CA9313343 |
74 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9313341 rs774346333 |
75 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747798983 CA9313342 |
75 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1003925988 CA306215412 |
76 | F>L | No |
ClinGen TOPMed gnomAD |
|
|
rs768588696 CA9313340 |
78 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA9313339 rs749027975 |
79 | A>D | No |
ClinGen ExAC gnomAD |
|
|
rs763584828 CA9313338 |
80 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA306215401 COSM1611837 rs368085900 |
80 | M>V | liver [Cosmic] | No |
ClinGen cosmic curated ESP TOPMed gnomAD |
|
CA404828916 rs1355349814 |
82 | P>T | No |
ClinGen TOPMed |
|
|
rs1366617087 CA404828880 |
83 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
CA404828898 rs1182452268 |
83 | E>K | No |
ClinGen gnomAD |
|
|
CA404828857 rs1568412140 |
84 | P>L | No |
ClinGen Ensembl |
|
|
CA404828864 rs756031889 |
84 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs756031889 CA9313337 |
84 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs374983500 CA9313335 |
85 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9313334 rs535000575 |
85 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs752925784 CA9313333 |
86 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA9313331 rs11574806 |
87 | A>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
VAR_044225 rs11574806 CA9313330 |
87 | A>V | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA9313329 rs552738926 |
88 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs772664956 CA9313327 |
89 | A>D | No |
ClinGen ExAC |
|
|
CA404828731 rs1312164985 |
90 | P>A | No |
ClinGen gnomAD |
|
|
CA9313326 rs767041599 |
90 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA404828601 rs1452794116 |
95 | D>G | No |
ClinGen gnomAD |
|
|
CA404828567 rs1363380054 |
96 | I>L | No |
ClinGen gnomAD |
|
|
CA306214813 rs750954282 |
100 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313309 rs750954282 |
100 | G>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404827516 rs1272152659 |
101 | L>M | No |
ClinGen TOPMed gnomAD |
|
|
CA306214810 rs199981346 |
103 | R>K | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
rs1226654646 CA404827432 |
106 | T>A | No |
ClinGen gnomAD |
|
|
CA9313306 rs564752007 |
106 | T>M | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs763890504 CA9313304 |
107 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404827391 rs762688348 |
108 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762688348 CA9313303 |
108 | V>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA306214804 rs1007540103 |
111 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1466166801 CA404827355 |
111 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA404827358 rs1466166801 |
111 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA9313301 COSM260946 rs756695076 |
114 | S>L | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs544946458 CA9313300 |
116 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9313299 rs776661768 |
116 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs139527778 CA9313297 |
117 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1255663317 CA404827194 |
119 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1195143967 CA404827188 |
119 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA404827176 rs1345718743 |
120 | G>D | No |
ClinGen gnomAD |
|
|
rs1230722079 CA404827150 |
123 | V>L | No |
ClinGen gnomAD |
|
|
CA9313294 rs749514182 |
124 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs556938614 CA306214789 |
125 | V>I | No |
ClinGen 1000Genomes TOPMed gnomAD |
|
|
CA9313291 rs750717633 |
127 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs781770914 CA9313290 |
129 | T>M | No |
ClinGen ExAC gnomAD |
|
|
COSM993122 rs1341140154 CA404827072 |
130 | S>L | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA9313286 rs757785461 |
136 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1178842404 CA404827013 |
136 | R>W | No |
ClinGen gnomAD |
|
|
CA404827006 rs149393037 |
137 | V>L | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA306214774 rs149393037 |
137 | V>M | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA9313285 rs543340984 |
141 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA404826951 rs543340984 |
141 | P>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9313283 rs759397510 |
144 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759397510 CA404826927 |
144 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404826904 rs1190624475 |
146 | T>A | No |
ClinGen TOPMed |
|
|
rs1369810398 CA404826879 |
148 | G>D | No |
ClinGen TOPMed |
|
|
rs1278016314 CA404826863 |
150 | C>G | No |
ClinGen gnomAD |
|
|
rs760604410 CA9313280 COSM3822354 |
152 | V>I | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1484753127 CA404826773 |
155 | A>S | No |
ClinGen gnomAD |
|
|
rs1484753127 CA404826776 |
155 | A>T | No |
ClinGen gnomAD |
|
|
CA404826764 rs1396839068 |
156 | L>V | No |
ClinGen TOPMed |
|
|
CA404826698 rs1333693604 |
161 | P>L | No |
ClinGen TOPMed |
|
|
CA404826704 rs1333693604 |
161 | P>Q | No |
ClinGen TOPMed |
|
|
rs758809393 CA9313251 |
163 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA9313252 rs758809393 |
163 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs778281150 CA9313249 |
165 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313244 rs750173945 |
167 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA9313245 rs755935067 |
167 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9313243 COSM993121 rs767544339 |
168 | T>M | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA9313241 rs200492048 |
170 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1199547679 CA404826597 |
171 | V>F | No |
ClinGen TOPMed |
|
|
CA9313240 rs765242501 |
172 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA9313239 rs369362313 |
174 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9313238 rs549708240 |
175 | S>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs773417154 CA9313235 |
176 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs761170150 CA9313236 |
176 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA404826505 rs1191487364 |
179 | Y>C | No |
ClinGen gnomAD |
|
|
rs748588052 CA9313233 |
180 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA9313232 rs188185089 |
181 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1271172262 CA404826233 |
185 | S>G | No |
ClinGen TOPMed |
|
|
CA404826215 rs1178964320 |
186 | P>T | No |
ClinGen gnomAD |
|
|
rs753944406 CA9313221 |
188 | I>F | No |
ClinGen ExAC gnomAD |
|
|
CA404826152 rs1199023048 |
189 | P>H | No |
ClinGen TOPMed |
|
|
CA404826160 rs1211485653 |
189 | P>S | No |
ClinGen gnomAD |
|
|
rs113307565 CA9313219 |
190 | P>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA9313218 rs373750416 |
190 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA404826139 rs373750416 |
190 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9313220 rs113307565 |
190 | P>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA9313216 rs762172427 |
191 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
CA9313214 rs769103996 |
192 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313213 rs201140645 |
192 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1416600175 CA404825996 |
199 | T>N | No |
ClinGen gnomAD |
|
|
CA404825991 rs1373384104 |
200 | L>V | No |
ClinGen gnomAD |
|
|
CA404825973 rs1172704687 |
201 | K>E | No |
ClinGen gnomAD |
|
|
CA9313212 rs201710065 |
202 | T>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1161591394 CA404825919 |
204 | V>A | No |
ClinGen gnomAD |
|
|
CA404825892 rs1249967430 |
206 | G>R | No |
ClinGen gnomAD |
|
|
rs757031781 CA9313209 |
209 | L>M | No |
ClinGen ExAC gnomAD |
|
|
rs757031781 CA9313208 |
209 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA306214422 rs2314668 |
210 | E>Q | No |
ClinGen Ensembl |
|
|
CA404825741 rs1600535313 |
213 | T>R | No |
ClinGen Ensembl |
|
|
rs777617707 CA9313206 |
216 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA404825662 rs1305501146 |
217 | R>L | No |
ClinGen TOPMed |
|
|
CA9313204 rs754038245 |
218 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404825635 rs1236381009 |
219 | A>V | No |
ClinGen gnomAD |
|
|
CA306214413 rs868552092 |
221 | A>T | No |
ClinGen Ensembl |
|
|
CA404825601 rs1315833295 |
222 | N>S | No |
ClinGen gnomAD |
|
|
rs1234593802 CA404825589 |
223 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA9313202 rs375406802 |
223 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1187276385 CA404825564 |
225 | R>W | No |
ClinGen TOPMed |
|
|
CA404825532 rs1600535208 |
227 | C>G | No |
ClinGen Ensembl |
|
|
CA9313199 rs762107899 |
228 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1386388002 CA404825433 |
233 | Q>H | No |
ClinGen TOPMed |
|
|
rs763448432 CA404825408 |
235 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs763448432 CA9313196 |
235 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs745339783 CA404825395 |
236 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404825400 rs1600535144 |
236 | D>G | No |
ClinGen Ensembl |
|
|
CA9313194 rs769294379 |
236 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs776165734 CA9313192 |
238 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs1600535122 CA404825359 |
239 | L>P | No |
ClinGen Ensembl |
|
|
rs1600535090 CA404825276 |
245 | D>E | No |
ClinGen Ensembl |
|
|
rs1287886403 CA404825294 |
245 | D>N | No |
ClinGen TOPMed |
|
|
rs962206697 CA306214392 |
251 | I>V | No |
ClinGen Ensembl |
|
|
CA9313183 rs367943562 |
252 | T>I | No |
ClinGen ESP ExAC gnomAD |
|
|
CA404825177 rs1381435007 |
253 | S>C | No |
ClinGen gnomAD |
|
|
CA404825176 rs1381435007 |
253 | S>F | No |
ClinGen gnomAD |
|
|
rs1293094092 CA404825181 |
253 | S>T | No |
ClinGen gnomAD |
|
|
rs751939678 CA9313181 |
255 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA9313157 rs765926771 |
258 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA9313156 rs760036022 |
262 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA9313155 rs773031578 |
264 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1236771017 CA404821990 |
265 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs761750949 CA9313153 |
266 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs978206524 CA306214239 |
266 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA306214241 rs371165569 |
266 | Q>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs749237572 CA9313149 |
279 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA404821730 rs1433232980 |
281 | S>A | No |
ClinGen gnomAD |
|
|
rs778014771 CA404821639 |
286 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778014771 CA9313145 |
286 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs11541424 CA306214230 |
287 | H>Y | No |
ClinGen Ensembl |
|
|
CA9313144 rs758585209 |
289 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404821560 rs758585209 |
289 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313143 rs753124091 |
289 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758585209 CA404821557 |
289 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313141 rs755238463 |
290 | A>V | No |
ClinGen ExAC |
|
|
CA404821504 rs1208530110 |
293 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA404821493 rs1485206868 |
295 | C>R | No |
ClinGen gnomAD |
|
|
CA404821459 rs1278231522 |
297 | L>R | No |
ClinGen gnomAD |
|
|
rs974927432 CA306214219 |
302 | Q>E | No |
ClinGen Ensembl |
|
|
rs1375177195 CA404821139 |
311 | R>C | No |
ClinGen gnomAD |
|
|
CA9313134 rs745659664 |
311 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA9313133 rs768513900 |
314 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA404844682 rs1243704968 |
320 | G>E | No |
ClinGen TOPMed |
|
|
CA404844686 rs1418468436 |
320 | G>W | No |
ClinGen gnomAD |
|
|
CA404844613 rs1255115965 |
323 | S>G | No |
ClinGen gnomAD |
|
|
CA9313103 rs780279721 |
323 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA306210978 rs1007979517 |
324 | E>K | No |
ClinGen gnomAD |
|
|
CA306210977 rs953957801 |
327 | P>H | No |
ClinGen gnomAD |
|
|
rs953957801 CA404844503 |
327 | P>L | No |
ClinGen gnomAD |
|
|
CA404844022 rs1183392310 |
339 | N>K | No |
ClinGen TOPMed |
|
|
rs751720062 CA9313070 |
341 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs751720062 CA9313069 |
341 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1455741381 CA404843739 |
344 | A>E | No |
ClinGen gnomAD |
|
|
rs187595132 CA9313067 |
344 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs144976335 CA9313066 |
347 | S>L | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1008953397 CA306210927 |
350 | V>L | No |
ClinGen TOPMed |
|
|
rs1463412229 CA404843588 |
351 | K>R | No |
ClinGen gnomAD |
|
|
CA404843574 rs1312010714 |
352 | K>E | No |
ClinGen TOPMed |
|
|
CA9313063 rs777128222 |
353 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771523688 CA404843511 |
354 | A>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313062 rs771523688 |
354 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1464064536 CA404843497 |
355 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA9313060 rs200336385 |
355 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313058 rs747644640 |
356 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA9313057 rs202202469 |
357 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA404843456 rs1380453817 |
358 | S>N | No |
ClinGen TOPMed |
|
|
CA9313056 rs754595106 |
359 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9313055 rs367694382 |
362 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs553830583 CA9313054 |
363 | L>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA404843362 rs1335030032 |
365 | R>Q | No |
ClinGen TOPMed |
|
|
CA9313052 rs750335116 |
365 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1600527051 CA404843292 |
370 | N>T | No |
ClinGen Ensembl |
|
|
CA9313050 rs762968551 |
373 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767452570 CA9313051 |
373 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752805932 CA9313049 |
375 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA9313048 rs765617173 |
378 | C>F | No |
ClinGen ExAC gnomAD |
|
|
rs960850880 CA306210923 |
379 | P>R | No |
ClinGen Ensembl |
|
|
rs1166337925 CA404843042 |
383 | A>T | No |
ClinGen gnomAD |
|
|
CA9313036 rs755894944 |
386 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404842888 rs755894944 |
386 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1485858150 CA404842861 |
387 | P>S | No |
ClinGen gnomAD |
|
|
rs1213223035 CA404842816 |
388 | W>S | No |
ClinGen gnomAD |
|
|
CA404842755 rs1600526455 |
390 | W>* | No |
ClinGen Ensembl |
|
|
rs781044099 CA9313034 |
392 | F>V | No |
ClinGen ExAC gnomAD |
|
|
CA9313033 rs757210930 |
394 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1314862907 CA404842583 |
395 | T>I | No |
ClinGen TOPMed |
|
|
rs1235147076 CA404842563 |
396 | A>V | No |
ClinGen TOPMed |
|
|
rs765416957 CA9313031 |
397 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765416957 CA404842552 |
397 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA404842515 rs1409635138 |
398 | A>V | No |
ClinGen gnomAD |
|
|
CA404842486 rs755038922 |
399 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA306210869 rs933276965 |
400 | G>E | No |
ClinGen TOPMed |
|
|
CA404842435 rs1412336554 |
401 | G>D | No |
ClinGen gnomAD |
|
|
CA9313027 rs760993884 |
409 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1181306975 CA404842262 |
409 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs919262421 CA306210867 |
412 | N>I | No |
ClinGen TOPMed |
No associated diseases with Q14318
5 regional properties for Q14318
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FKBP-type peptidyl-prolyl cis-trans isomerase domain | 115 - 204 | IPR001179 |
| repeat | Tetratricopeptide repeat 2 | 307 - 339 | IPR013105 |
| repeat | Tetratricopeptide repeat | 228 - 261 | IPR019734-1 |
| repeat | Tetratricopeptide repeat | 272 - 305 | IPR019734-2 |
| repeat | Tetratricopeptide repeat | 306 - 339 | IPR019734-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 5.2.1.8 | Cis-trans isomerases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR46512 | PEPTIDYLPROLYL ISOMERASE |
| PANTHER Subfamily | PTHR46512:SF3 | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FKBP8 |
| PANTHER Protein Class | chaperone | |
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| integral component of endoplasmic reticulum membrane | The component of the endoplasmic reticulum membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| mitochondrial membrane | Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| disordered domain specific binding | Binding to a disordered domain of a protein. |
| identical protein binding | Binding to an identical protein or proteins. |
| metal ion binding | Binding to a metal ion. |
| peptidyl-prolyl cis-trans isomerase activity | Catalysis of the reaction: peptidyl-proline (omega=180) = peptidyl-proline (omega=0). |
| protein folding chaperone | Binding to a protein or a protein-containing complex to assist the protein folding process. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| camera-type eye development | The process whose specific outcome is the progression of the camera-type eye over time, from its formation to the mature structure. The camera-type eye is an organ of sight that receives light through an aperture and focuses it through a lens, projecting it on a photoreceptor field. |
| cell fate specification | The process involved in the specification of cell identity. Once specification has taken place, a cell will be committed to differentiate down a specific pathway if left in its normal environment. |
| dorsal/ventral neural tube patterning | The process in which the neural tube is regionalized in the dorsoventral axis. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| multicellular organism growth | The increase in size or mass of an entire multicellular organism, as opposed to cell growth. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of protein phosphorylation | Any process that stops, prevents or reduces the rate of addition of phosphate groups to amino acids within a protein. |
| positive regulation of BMP signaling pathway | Any process that activates or increases the frequency, rate or extent of BMP signaling pathway activity. |
| protein folding | The process of assisting in the covalent and noncovalent assembly of single chain polypeptides or multisubunit complexes into the correct tertiary structure. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| smoothened signaling pathway | The series of molecular signals generated as a consequence of activation of the transmembrane protein Smoothened. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A6QQ71 | FKBP6 | Inactive peptidyl-prolyl cis-trans isomerase FKBP6 | Bos taurus (Bovine) | PR |
| Q9W1I9 | shu | Inactive peptidyl-prolyl cis-trans isomerase shutdown | Drosophila melanogaster (Fruit fly) | PR |
| Q02790 | FKBP4 | Peptidyl-prolyl cis-trans isomerase FKBP4 | Homo sapiens (Human) | PR |
| O35465 | Fkbp8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Mus musculus (Mouse) | SS |
| Q3B7U9 | Fkbp8 | Peptidyl-prolyl cis-trans isomerase FKBP8 | Rattus norvegicus (Rat) | SS |
| Q7DMA9 | PAS1 | Peptidyl-prolyl cis-trans isomerase PASTICCINO1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASCAEPSEP | SAPLPAGVPP | LEDFEVLDGV | EDAEGEEEEE | EEEEEEDDLS | ELPPLEDMGQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PPAEEAEQPG | ALAREFLAAM | EPEPAPAPAP | EEWLDILGNG | LLRKKTLVPG | PPGSSRPVKG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| QVVTVHLQTS | LENGTRVQEE | PELVFTLGDC | DVIQALDLSV | PLMDVGETAM | VTADSKYCYG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PQGRSPYIPP | HAALCLEVTL | KTAVDGPDLE | MLTGQERVAL | ANRKRECGNA | HYQRADFVLA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ANSYDLAIKA | ITSSAKVDMT | FEEEAQLLQL | KVKCLNNLAA | SQLKLDHYRA | ALRSCSLVLE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HQPDNIKALF | RKGKVLAQQG | EYSEAIPILR | AALKLEPSNK | TIHAELSKLV | KKHAAQRSTE |
| 370 | 380 | 390 | 400 | 410 | |
| TALYRKMLGN | PSRLPAKCPG | KGAWSIPWKW | LFGATAVALG | GVALSVVIAA | RN |