Q14240
Gene name |
EIF4A2 (DDX2B, EIF4F) |
Protein name |
Eukaryotic initiation factor 4A-II |
Names |
eIF-4A-II, eIF4A-II, ATP-dependent RNA helicase eIF4A-2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1974 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
|
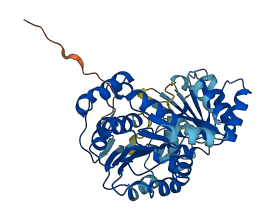
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q14240
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3BOR | X-ray | 185 A | A | 22-240 | PDB |
| AF-Q14240-F1 | Predicted | AlphaFoldDB |
149 variants for Q14240
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA2746667 COSM400629 rs764605841 |
2 | S>C | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs764605841 CA2746666 |
2 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs760934867 CA2746665 |
2 | S>T | No |
ClinGen ExAC |
|
|
CA2746669 COSM77639 rs765785108 |
3 | G>V | ovary [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA355719308 rs960682898 |
4 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs750826078 CA2746670 |
4 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs960682898 CA89648401 |
4 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
CA355719313 rs1560082688 |
5 | S>A | No |
ClinGen Ensembl |
|
|
CA2746672 rs779490566 |
5 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355719322 rs779490566 COSM420090 |
5 | S>F | Variant assessed as Somatic; impact. urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs754593091 CA89648439 |
6 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754593091 CA2746674 |
6 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA355719343 rs1423569431 |
7 | D>E | No |
ClinGen gnomAD |
|
|
rs1560082706 CA355719340 |
7 | D>G | No |
ClinGen Ensembl |
|
|
rs374075692 CA2746675 |
7 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs908631706 CA89648440 |
8 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1377442083 CA355719358 |
9 | N>D | No |
ClinGen gnomAD |
|
|
CA2746677 rs143326381 |
9 | N>K | No |
ClinGen ESP ExAC |
|
|
CA355719364 rs1418086849 |
9 | N>S | No |
ClinGen gnomAD |
|
|
rs1418086849 CA355719363 |
9 | N>T | No |
ClinGen gnomAD |
|
|
CA355719370 rs1317806581 |
10 | R>G | No |
ClinGen gnomAD |
|
|
rs1479988881 CA355719374 |
10 | R>T | No |
ClinGen TOPMed |
|
|
rs1553797619 CA2746717 |
12 | H>Q | No |
ClinGen Ensembl |
|
|
CA2746719 rs753573326 |
15 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757324676 CA2746720 |
16 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA355720469 rs1381743588 |
18 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA2746721 rs778995582 |
20 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs371691662 | 26 | S>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1169693407 CA355720551 |
26 | S>T | No |
ClinGen TOPMed |
|
|
CA2746753 rs187041507 |
27 | N>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA89650365 rs17851348 |
27 | N>S | No |
ClinGen TOPMed |
|
| TCGA novel | 30 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355720631 rs953593626 |
37 | D>N | No |
ClinGen gnomAD |
|
|
rs1248318962 CA355720635 |
37 | D>V | No |
ClinGen TOPMed |
|
|
CA89650389 rs953593626 |
37 | D>Y | No |
ClinGen gnomAD |
|
|
rs1382097286 CA355720683 |
43 | S>F | No |
ClinGen gnomAD |
|
|
rs201474676 CA89650421 |
45 | L>F | No |
ClinGen 1000Genomes TOPMed |
|
|
rs867137018 CA89650442 |
47 | G>D | No |
ClinGen Ensembl |
|
|
rs11538618 CA89650451 |
48 | I>S | No |
ClinGen Ensembl |
|
|
CA89650449 rs972376732 |
48 | I>V | No |
ClinGen gnomAD |
|
|
CA355720716 rs1328591615 |
49 | Y>C | No |
ClinGen gnomAD |
|
|
rs200752693 CA89650482 |
55 | K>E | No |
ClinGen 1000Genomes |
|
|
CA355720785 rs1560083657 |
58 | A>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
| TCGA novel | 58 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2746762 rs776167777 |
58 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA89650504 rs577099369 |
63 | A>G | No |
ClinGen 1000Genomes TOPMed |
|
| TCGA novel | 63 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355720855 rs577099369 |
63 | A>V | No |
ClinGen 1000Genomes TOPMed |
|
|
CA2746765 rs765159897 |
64 | I>V | No |
ClinGen ExAC |
|
|
rs750238082 CA2746766 |
65 | I>L | No |
ClinGen ExAC gnomAD |
|
| rs755185062 | 65 | I>missing | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA89650510 rs866637500 |
66 | P>L | No |
ClinGen Ensembl |
|
|
rs1480206121 CA355721036 |
73 | V>M | No |
ClinGen gnomAD |
|
|
rs11538611 CA89651048 |
76 | Q>R | No |
ClinGen Ensembl |
|
| TCGA novel | 77 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2746839 rs773335544 |
81 | T>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 83 | K>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355721237 rs1579157436 |
84 | T>P | No |
ClinGen Ensembl |
|
|
rs11538614 CA89651060 |
85 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA89651066 rs775555447 |
86 | T>S | No |
ClinGen Ensembl |
|
|
rs201339295 CA89651089 |
92 | L>P | No |
ClinGen 1000Genomes |
|
|
VAR_052158 CA89651090 rs11538616 |
93 | Q>H | No |
ClinGen UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs751663411 CA2746842 |
94 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs755425078 CA355721416 |
97 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1368492875 CA355721411 |
97 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA355721424 rs1579157500 |
98 | E>D | No |
ClinGen Ensembl |
|
|
CA355721426 rs1579157504 |
99 | F>L | No |
ClinGen Ensembl |
|
|
CA355721437 rs1366744628 |
100 | K>T | No |
ClinGen TOPMed |
|
|
rs753306564 CA2746845 |
103 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1050836155 CA89651142 |
109 | P>T | No |
ClinGen Ensembl |
|
|
CA355721557 rs1560084030 |
112 | E>Q | No |
ClinGen Ensembl |
|
|
rs1407908695 CA355722036 |
128 | M>T | No |
ClinGen gnomAD |
|
|
CA355722060 rs1327710742 |
129 | G>E | No |
ClinGen TOPMed gnomAD |
|
|
CA355722064 rs1327710742 |
129 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
CA2746872 rs756828110 |
131 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA89652095 rs991933918 |
134 | A>V | No |
ClinGen Ensembl |
|
| TCGA novel | 136 | I>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355722270 rs1579159855 |
140 | N>S | No |
ClinGen Ensembl |
|
|
CA2746875 rs758037831 |
143 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 147 | K>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355722401 rs1195471558 |
150 | A>T | No |
ClinGen gnomAD |
|
| TCGA novel | 152 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355722455 rs1560084659 |
154 | H>R | No |
ClinGen Ensembl |
|
|
CA89652103 rs11538613 |
154 | H>Y | No |
ClinGen Ensembl |
|
|
CA2746879 rs142804104 |
155 | I>T | No |
ClinGen ESP ExAC |
|
|
rs746817318 CA2746878 |
155 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| rs767682413 | 155 | I>missing | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2746880 COSM730070 rs781218973 |
156 | V>I | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA2746884 rs774265072 CA2746883 |
167 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 174 | P>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 176 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2746925 rs754554886 |
176 | W>G | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 179 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 179 | M>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_035838 | 181 | V>L | a breast cancer sample; somatic mutation [UniProt] | No | UniProt |
| TCGA novel | 181 | V>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 183 | D>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 184 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 185 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 194 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1248522662 CA355723209 |
195 | D>V | No |
ClinGen TOPMed |
|
| TCGA novel | 197 | I>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 202 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2746932 rs749201932 |
207 | S>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 208 | I>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs765266132 CA2746971 |
219 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA2746972 rs750332310 |
219 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA355723570 rs1309985225 |
223 | E>K | No |
ClinGen gnomAD |
|
|
CA2746973 rs755032307 |
233 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs868809630 CA89654935 |
248 | K>R | No |
ClinGen Ensembl |
|
| TCGA novel | 251 | Y>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355724241 rs1299766512 |
256 | R>K | No |
ClinGen TOPMed |
|
| TCGA novel | 265 | C>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355724446 rs1170283376 |
265 | C>S | No |
ClinGen gnomAD |
|
|
rs1237593933 CA355724465 |
267 | L>S | No |
ClinGen TOPMed |
|
|
rs79277600 CA2747088 |
277 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1579163623 CA355724608 |
280 | L>F | No |
ClinGen Ensembl |
|
|
rs1980299 CA89655985 |
281 | N>K | No |
ClinGen Ensembl |
|
|
CA355725053 rs1579163654 |
286 | V>G | No |
ClinGen Ensembl |
|
|
CA355725125 rs1579163666 |
290 | T>A | No |
ClinGen Ensembl |
|
|
rs1221692747 CA355725167 |
291 | E>D | No |
ClinGen TOPMed |
|
|
rs754199367 CA89656016 |
291 | E>G | No |
ClinGen Ensembl |
|
| TCGA novel | 301 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA89656079 rs1045274054 |
302 | A>S | No |
ClinGen TOPMed |
|
| TCGA novel | 306 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1579164782 CA355725744 |
311 | E>D | No |
ClinGen Ensembl |
|
|
CA2747171 rs748724172 |
313 | D>E | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 342 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA2747245 rs769438133 |
344 | L>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 347 | N>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs762856383 CA2747247 |
352 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA355727394 rs1213509261 |
366 | R>* | No |
ClinGen TOPMed |
|
| TCGA novel | 366 | R>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 366 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776557195 CA2747294 |
373 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA355727721 rs1560087652 |
379 | E>G | No |
ClinGen Ensembl |
|
| TCGA novel | 379 | E>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 381 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 382 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA355727909 rs1427878661 COSM1042378 |
386 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA89658455 rs866118494 |
390 | T>N | No |
ClinGen Ensembl |
|
|
CA89658458 rs554887819 |
392 | Y>C | No |
ClinGen 1000Genomes |
|
|
CA355728121 rs1335839025 |
394 | T>A | No |
ClinGen gnomAD |
|
|
rs1406419091 CA355728139 |
395 | T>A | No |
ClinGen gnomAD |
|
|
CA89658462 rs199556041 COSM1484942 |
395 | T>I | Variant assessed as Somatic; impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
CA89658460 rs199556041 |
395 | T>R | No |
ClinGen TOPMed gnomAD |
|
|
CA917088590 rs1579171264 |
403 | V>* | No |
ClinGen Ensembl |
|
|
rs35049134 CA89658497 |
405 | D>A | No |
ClinGen Ensembl |
|
| TCGA novel | 407 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q14240
1 regional properties for Q14240
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | TATA-box binding protein, conserved site | 295 - 344 | IPR030491 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| eukaryotic translation initiation factor 4F complex | The eukaryotic translation initiation factor 4F complex is composed of eIF4E, eIF4A and eIF4G; it is involved in the recognition of the mRNA cap, ATP-dependent unwinding of the 5'-terminal secondary structure and recruitment of the mRNA to the ribosome. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| helicase activity | Catalysis of the reaction: ATP + H2O = ADP + phosphate, to drive the unwinding of a DNA or RNA helix. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
| translation initiation factor activity | Functions in the initiation of ribosome-mediated translation of mRNA into a polypeptide. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to leukemia inhibitory factor | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a leukemia inhibitory factor stimulus. |
| cytoplasmic translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein in the cytoplasm. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
| negative regulation of RNA-directed 5'-3' RNA polymerase activity | Any process that stops, prevents or reduces the frequency, rate or extent of RNA-directed 5'-3' RNA polymerase activity. |
| regulation of translational initiation | Any process that modulates the frequency, rate or extent of translational initiation. |
| translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P41378 | Eukaryotic initiation factor 4A | Triticum aestivum (Wheat) | PR | |
| P10081 | TIF2 | ATP-dependent RNA helicase eIF4A | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q3SZ54 | EIF4A1 | Eukaryotic initiation factor 4A-I | Bos taurus (Bovine) | PR |
| Q3SZ65 | EIF4A2 | Eukaryotic initiation factor 4A-II | Bos taurus (Bovine) | PR |
| Q8JFP1 | EIF4A2 | Eukaryotic initiation factor 4A-II | Gallus gallus (Chicken) | PR |
| A5A6N4 | EIF4A1 | Eukaryotic initiation factor 4A-I | Pan troglodytes (Chimpanzee) | PR |
| Q8NHQ9 | DDX55 | ATP-dependent RNA helicase DDX55 | Homo sapiens (Human) | PR |
| P60842 | EIF4A1 | Eukaryotic initiation factor 4A-I | Homo sapiens (Human) | PR |
| Q9Y6V7 | DDX49 | Probable ATP-dependent RNA helicase DDX49 | Homo sapiens (Human) | PR |
| Q9H0S4 | DDX47 | Probable ATP-dependent RNA helicase DDX47 | Homo sapiens (Human) | PR |
| Q8TDD1 | DDX54 | ATP-dependent RNA helicase DDX54 | Homo sapiens (Human) | PR |
| Q9GZR7 | DDX24 | ATP-dependent RNA helicase DDX24 | Homo sapiens (Human) | PR |
| Q92499 | DDX1 | ATP-dependent RNA helicase DDX1 | Homo sapiens (Human) | PR |
| Q9NY93 | DDX56 | Probable ATP-dependent RNA helicase DDX56 | Homo sapiens (Human) | PR |
| Q9NUL7 | DDX28 | Probable ATP-dependent RNA helicase DDX28 | Homo sapiens (Human) | PR |
| Q7L014 | DDX46 | Probable ATP-dependent RNA helicase DDX46 | Homo sapiens (Human) | PR |
| Q41741 | Eukaryotic initiation factor 4A | Zea mays (Maize) | PR | |
| P60843 | Eif4a1 | Eukaryotic initiation factor 4A-I | Mus musculus (Mouse) | PR |
| P10630 | Eif4a2 | Eukaryotic initiation factor 4A-II | Mus musculus (Mouse) | PR |
| Q5RKI1 | Eif4a2 | Eukaryotic initiation factor 4A-II | Rattus norvegicus (Rat) | PR |
| Q6Z2Z4 | Os02g0146600 | Eukaryotic initiation factor 4A-3 | Oryza sativa subsp japonica (Rice) | PR |
| P35683 | Os06g0701100 | Eukaryotic initiation factor 4A-1 | Oryza sativa subsp japonica (Rice) | PR |
| P27639 | inf-1 | Eukaryotic initiation factor 4A | Caenorhabditis elegans | PR |
| Q3E9C3 | RH58 | DEAD-box ATP-dependent RNA helicase 58, chloroplastic | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9CAI7 | TIF4A-3 | Eukaryotic initiation factor 4A-3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41376 | EIF4A1 | Eukaryotic initiation factor 4A-1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P41377 | TIF4A-2 | Eukaryotic initiation factor 4A-2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGGSADYNR | EHGGPEGMDP | DGVIESNWNE | IVDNFDDMNL | KESLLRGIYA | YGFEKPSAIQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QRAIIPCIKG | YDVIAQAQSG | TGKTATFAIS | ILQQLEIEFK | ETQALVLAPT | RELAQQIQKV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ILALGDYMGA | TCHACIGGTN | VRNEMQKLQA | EAPHIVVGTP | GRVFDMLNRR | YLSPKWIKMF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VLDEADEMLS | RGFKDQIYEI | FQKLNTSIQV | VLLSATMPTD | VLEVTKKFMR | DPIRILVKKE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ELTLEGIKQF | YINVEREEWK | LDTLCDLYET | LTITQAVIFL | NTRRKVDWLT | EKMHARDFTV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SALHGDMDQK | ERDVIMREFR | SGSSRVLITT | DLLARGIDVQ | QVSLVINYDL | PTNRENYIHR |
| 370 | 380 | 390 | 400 | ||
| IGRGGRFGRK | GVAINFVTEE | DKRILRDIET | FYNTTVEEMP | MNVADLI |