Q14168
Gene name |
MPP2 |
Protein name |
MAGUK p55 subfamily member 2 |
Names |
Discs large homolog 2, Protein MPP2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4355 |
EC number |
|
Protein Class |
|
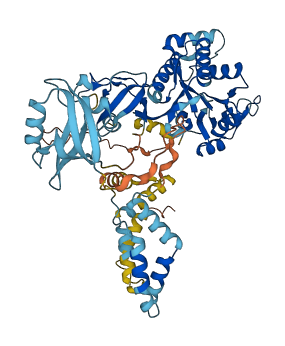
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for Q14168
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2E7K | NMR | - | A | 163-240 | PDB |
| AF-Q14168-F1 | Predicted | AlphaFoldDB |
No variants for Q14168
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q14168 | |||||
No associated diseases with Q14168
Functions
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoskeleton | A cellular structure that forms the internal framework of eukaryotic and prokaryotic cells. The cytoskeleton includes intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles. |
| dendrite membrane | The portion of the plasma membrane surrounding a dendrite. |
| dendritic shaft | Cylindric portion of the dendrite, directly stemming from the perikaryon, and carrying the dendritic spines. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| excitatory postsynaptic potential | A process that leads to a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| long-term synaptic potentiation | A process that modulates synaptic plasticity such that synapses are changed resulting in the increase in the rate, or frequency of synaptic transmission at the synapse. |
| protein homooligomerization | The process of creating protein oligomers, compounds composed of a small number, usually between three and ten, of identical component monomers. Oligomers may be formed by the polymerization of a number of monomers or the depolymerization of a large protein polymer. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q24210 | CASK | Peripheral plasma membrane protein CASK | Drosophila melanogaster (Fruit fly) | SS |
| Q00013 | MPP1 | 55 kDa erythrocyte membrane protein | Homo sapiens (Human) | PR |
| O14936 | CASK | Peripheral plasma membrane protein CASK | Homo sapiens (Human) | EV |
| Q9NZW5 | PALS2 | Protein PALS2 | Homo sapiens (Human) | PR |
| Q96JB8 | MPP4 | MAGUK p55 subfamily member 4 | Homo sapiens (Human) | PR |
| O70589 | Cask | Peripheral plasma membrane protein CASK | Mus musculus (Mouse) | SS |
| P70290 | Mpp1 | 55 kDa erythrocyte membrane protein | Mus musculus (Mouse) | PR |
| O88910 | Mpp3 | MAGUK p55 subfamily member 3 | Mus musculus (Mouse) | PR |
| Q6P7F1 | Mpp4 | MAGUK p55 subfamily member 4 | Mus musculus (Mouse) | PR |
| Q8BVD5 | Mpp7 | MAGUK p55 subfamily member 7 | Mus musculus (Mouse) | PR |
| Q9JLB0 | Pals2 | Protein PALS2 | Mus musculus (Mouse) | PR |
| Q9WV34 | Mpp2 | MAGUK p55 subfamily member 2 | Mus musculus (Mouse) | PR |
| Q9QYH1 | Mpp4 | MAGUK p55 subfamily member 4 | Rattus norvegicus (Rat) | PR |
| P54936 | lin-2 | Protein lin-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPVAATNSET | AMQQVLDNLG | SLPSATGAAE | LDLIFLRGIM | ESPIVRSLAK | VIMVLWFMQQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NVFVPMKYML | KYFGAHERLE | ETKLEAVRDN | NLELVQEILR | DLAHVAEQSS | TAAELAHILQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EPHFQSLLET | HDSVASKTYE | TPPPSPGLDP | TFSNQPVPPD | AVRMVGIRKT | AGEHLGVTFR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VEGGELVIAR | ILHGGMVAQQ | GLLHVGDIIK | EVNGQPVGSD | PRALQELLRN | ASGSVILKIL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PSYQEPHLPR | QVFVKCHFDY | DPARDSLIPC | KEAGLRFNAG | DLLQIVNQDD | ANWWQACHVE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GGSAGLIPSQ | LLEEKRKAFV | KRDLELTPNS | GTLCGSLSGK | KKKRMMYLTT | KNAEFDRHEL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LIYEEVARMP | PFRRKTLVLI | GAQGVGRRSL | KNKLIMWDPD | RYGTTVPYTS | RRPKDSEREG |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QGYSFVSRGE | MEADVRAGRY | LEHGEYEGNL | YGTRIDSIRG | VVAAGKVCVL | DVNPQAVKVL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| RTAEFVPYVV | FIEAPDFETL | RAMNRAALES | GISTKQLTEA | DLRRTVEESS | RIQRGYGHYF |
| 550 | 560 | 570 | |||
| DLCLVNSNLE | RTFRELQTAM | EKLRTEPQWV | PVSWVY |