Q13188
Gene name |
STK3 (KRS1, MST2) |
Protein name |
Serine/threonine-protein kinase 3 |
Names |
Mammalian STE20-like protein kinase 2, MST-2, STE20-like kinase MST2, Serine/threonine-protein kinase Krs-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6788 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
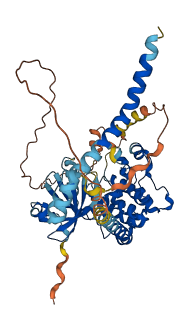
Descriptions
The Mst1/2 kinases are cytosolic Ste20-related kinases activated by autophosphorylation. Designated class II GC kinases, Mst1 and Mst2, share 76% sequence identity and contain an N-terminal catalytic domain, followed successively by an autoinhibitory segment and a coiled-coil SARAH domain that mediates hetero- and homo-dimerisation. Mst1 and Mst2 also become activated in cells undergoing apoptosis from a variety of stimuli. The activation mechanism is unclear, however, once activated, both kinases undergo cleavage by caspase3 at sites just carboxy terminal to their catalytic domains. The resultant catalytic polypeptides display an altered substrate specificity and lack their autoinhibitory domain. The caspase-cleaved catalytic fragments are highly and constitutively active with unrestricted nuclear access.
Autoinhibitory domains (AIDs)
Target domain |
27-278 (Protein kinase domain) |
Relief mechanism |
Cleavage |
Assay |
|
Accessory elements
163-186 (Activation loop from InterPro)
Target domain |
27-278 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Avruch J et al. (2011) "Mst1/2 signalling to Yap: gatekeeper for liver size and tumour development", British journal of cancer, 104, 24-32
- Zhou D et al. (2011) "Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance", Proceedings of the National Academy of Sciences of the United States of America, 108, E1312-20
- Creasy CL et al. (1996) "The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain", The Journal of biological chemistry, 271, 21049-53
Autoinhibited structure

Activated structure

12 structures for Q13188
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3WWS | X-ray | 201 A | A/B/C/D | 436-484 | PDB |
| 4HKD | X-ray | 150 A | A/B/C/D | 436-484 | PDB |
| 4L0N | X-ray | 140 A | A/B/C/D/E/F/G/H/I/J | 436-484 | PDB |
| 4LG4 | X-ray | 242 A | A/B/C/D/E/F | 16-313 | PDB |
| 4LGD | X-ray | 305 A | A/B/C/D | 9-491 | PDB |
| 4OH9 | X-ray | 170 A | A/B | 436-484 | PDB |
| 5BRM | X-ray | 265 A | G/H/I/J/K/L/M/N/O | 371-401 | PDB |
| 5DH3 | X-ray | 247 A | A/B | 15-313 | PDB |
| 6AO5 | X-ray | 296 A | A | 16-491 | PDB |
| 6AR2 | X-ray | 155 A | C/D | 373-382 | PDB |
| 8A66 | X-ray | 190 A | A/B | 16-312 | PDB |
| AF-Q13188-F1 | Predicted | AlphaFoldDB |
311 variants for Q13188
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA371854923 rs1265591568 |
2 | E>D | No |
ClinGen gnomAD |
|
|
CA371854914 rs1220390079 |
3 | Q>H | No |
ClinGen gnomAD |
|
|
CA371854908 rs1356943297 |
4 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1033189718 CA182835904 |
6 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1376045996 CA371854891 |
7 | P>L | No |
ClinGen gnomAD |
|
|
CA371854894 rs1240650026 |
7 | P>S | No |
ClinGen gnomAD |
|
|
CA371854879 rs1340266195 |
9 | S>N | No |
ClinGen TOPMed |
|
|
CA371855139 rs1157289261 |
9 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
rs746632419 CA4821992 |
14 | L>V | No |
ClinGen ExAC |
|
|
rs1378759384 CA371855084 |
17 | D>G | No |
ClinGen gnomAD |
|
|
CA371855065 rs1473562696 |
20 | T>A | No |
ClinGen TOPMed |
|
|
CA371855006 rs1426866722 |
28 | D>N | No |
ClinGen gnomAD |
|
|
CA371854982 rs1193956217 |
31 | E>G | No |
ClinGen gnomAD |
|
|
rs1487811766 CA371854978 |
32 | K>Q | No |
ClinGen gnomAD |
|
|
rs1202010149 CA371854957 |
35 | E>K | No |
ClinGen gnomAD |
|
|
CA4821968 rs187757501 |
40 | S>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4821967 rs745606479 |
44 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1403140497 CA371854778 |
49 | S>T | No |
ClinGen gnomAD |
|
|
rs749191480 CA4821964 |
50 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs762429515 CA182822632 |
51 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1164638081 CA371854752 |
53 | V>F | No |
ClinGen gnomAD |
|
|
rs1164638081 CA371854754 |
53 | V>I | No |
ClinGen gnomAD |
|
|
rs756129319 CA4821962 |
54 | A>G | No |
ClinGen ExAC |
|
|
CA371854748 rs1384400929 |
54 | A>S | No |
ClinGen gnomAD |
|
|
CA4821960 rs752970275 |
56 | K>Q | No |
ClinGen ExAC |
|
|
rs1228869369 CA371854727 |
57 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
rs1228869369 CA371854728 |
57 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
CA371854699 rs1356927652 |
61 | E>G | No |
ClinGen TOPMed |
|
|
CA371854695 rs1450868524 |
62 | S>T | No |
ClinGen TOPMed |
|
|
rs1345272410 CA371854646 |
68 | I>M | No |
ClinGen gnomAD |
|
|
CA4821957 rs751921239 |
70 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1226602594 CA371854598 |
75 | Q>* | No |
ClinGen TOPMed |
|
|
rs77105316 CA182822618 |
77 | C>S | No |
ClinGen Ensembl |
|
|
rs1268524562 CA371853775 |
80 | P>S | No |
ClinGen gnomAD |
|
|
CA371853757 rs1563960978 |
83 | V>I | No |
ClinGen Ensembl |
|
|
CA182818484 rs906166047 |
85 | Y>C | No |
ClinGen Ensembl |
|
|
CA4821932 rs200521127 |
93 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4821930 rs528318298 |
98 | V>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs947958279 CA371853602 |
104 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
rs947958279 CA182818475 |
104 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs773157841 CA4821927 |
105 | G>C | No |
ClinGen ExAC gnomAD |
|
|
rs1398850890 CA371853588 |
106 | S>C | No |
ClinGen gnomAD |
|
|
rs1350908369 CA371853571 |
109 | D>G | No |
ClinGen TOPMed |
|
|
CA182818470 rs920451180 |
110 | I>M | No |
ClinGen Ensembl |
|
|
CA371853563 rs1427248002 |
110 | I>T | No |
ClinGen gnomAD |
|
|
rs973207295 CA182818468 |
111 | I>V | No |
ClinGen Ensembl |
|
|
CA182818466 rs971193348 |
113 | L>* | No |
ClinGen TOPMed gnomAD |
|
|
CA4821924 COSM1726519 rs781199889 |
114 | R>* | liver [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA182818461 rs941475589 |
114 | R>L | No |
ClinGen TOPMed gnomAD |
|
|
COSM1458843 rs941475589 CA182818463 |
114 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA4821923 rs768771687 |
115 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA4821921 rs780352961 |
117 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1255925875 CA371854537 |
119 | I>M | No |
ClinGen TOPMed |
|
|
rs577272361 CA4821863 |
121 | D>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs371706117 CA371854513 |
123 | I>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs371706117 CA4821862 |
123 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1198258205 CA371854504 |
124 | A>E | No |
ClinGen gnomAD |
|
|
rs1429473482 CA371854507 |
124 | A>T | No |
ClinGen gnomAD |
|
|
rs1199182127 CA371854499 |
125 | T>A | No |
ClinGen TOPMed |
|
|
rs763763606 CA4821861 |
125 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1205550787 CA371854495 |
126 | I>V | No |
ClinGen gnomAD |
|
|
CA182795484 rs538970252 |
131 | L>S | No |
ClinGen gnomAD |
|
|
rs767717884 CA4821858 |
132 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1554657136 CA371854421 |
137 | L>* | No |
ClinGen Ensembl |
|
|
rs774457582 CA4821856 |
138 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371854395 rs1298163017 |
140 | M>I | No |
ClinGen gnomAD |
|
|
rs575295558 CA4821855 |
140 | M>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1383186928 CA371854393 |
141 | R>G | No |
ClinGen gnomAD |
|
|
rs367744890 CA4821854 |
143 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA371854349 rs1473249180 |
147 | I>V | No |
ClinGen TOPMed |
|
|
rs770128448 CA4821852 |
149 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1406203153 CA371854322 |
151 | N>D | No |
ClinGen gnomAD |
|
|
rs1438380177 CA371854309 |
152 | I>M | No |
ClinGen Ensembl |
|
|
CA371854292 rs1193626574 |
155 | N>S | No |
ClinGen gnomAD |
|
|
rs371776932 CA4821849 |
156 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371776932 CA4821848 |
156 | T>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs753429894 CA4821845 |
159 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371854255 rs1203171424 |
161 | K>Q | No |
ClinGen gnomAD |
|
|
CA371854207 rs1275978741 |
168 | A>P | No |
ClinGen TOPMed |
|
|
rs755916185 CA4821843 |
170 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA371854187 rs1587367870 |
171 | L>V | No |
ClinGen Ensembl |
|
|
CA4821842 rs752687054 |
172 | T>K | No |
ClinGen ExAC |
|
|
CA182795434 rs940407987 |
173 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
CA4821824 rs777502225 |
176 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA182795433 rs913508602 COSM1102929 |
178 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1587366532 CA371854129 |
178 | R>S | No |
ClinGen Ensembl |
|
|
rs1393938290 CA371854120 |
179 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA371854111 rs1302370737 |
180 | T>I | No |
ClinGen TOPMed |
|
|
CA371854100 rs1165228727 |
182 | I>T | No |
ClinGen gnomAD |
|
|
CA371854087 rs1587366422 |
184 | T>N | No |
ClinGen Ensembl |
|
|
CA4821822 rs752588921 |
188 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1344357232 CA371854047 |
190 | P>A | No |
ClinGen gnomAD |
|
|
rs781102717 CA4821821 |
192 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs992802416 CA182795429 |
192 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1174551470 CA371854025 |
193 | I>S | No |
ClinGen Ensembl |
|
|
CA371854015 rs1365638423 |
195 | E>K | No |
ClinGen gnomAD |
|
|
CA4821820 rs755030771 |
196 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs1402037240 CA371853999 |
197 | G>D | No |
ClinGen gnomAD |
|
|
rs960039884 CA182795427 |
197 | G>S | No |
ClinGen Ensembl |
|
|
rs775418746 CA182795426 |
198 | Y>C | No |
ClinGen TOPMed |
|
|
rs1182372085 CA371853980 |
200 | C>S | No |
ClinGen TOPMed gnomAD |
|
|
CA371853948 rs1262074313 |
204 | I>M | No |
ClinGen TOPMed |
|
|
CA4821818 rs766490046 |
204 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762870573 CA4821817 |
205 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA371853946 rs1476706609 |
205 | W>R | No |
ClinGen TOPMed |
|
|
CA4821816 rs750684813 |
207 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs765399291 CA4821815 |
208 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA182795424 rs952868146 |
209 | I>V | No |
ClinGen gnomAD |
|
|
rs1237780141 CA371853901 |
212 | I>T | No |
ClinGen gnomAD |
|
|
CA4821812 rs764439932 |
214 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1424205963 CA371853890 |
214 | M>V | No |
ClinGen TOPMed |
|
|
CA4821811 rs761304300 |
215 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371853870 rs1405273639 |
216 | E>D | No |
ClinGen gnomAD |
|
|
rs775804009 CA4821810 |
218 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA4821809 rs779203218 |
219 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1408691717 CA371853849 |
220 | P>S | No |
ClinGen gnomAD |
|
|
CA4821808 rs748708313 |
224 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA4821807 rs564202745 |
228 | R>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs769609076 CA4821806 |
228 | R>M | No |
ClinGen ExAC gnomAD |
|
|
CA371853506 rs1357369489 |
229 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA4821787 rs776353449 |
235 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371853449 rs1586967148 |
237 | P>A | No |
ClinGen Ensembl |
|
|
CA371853440 rs1216047422 |
238 | P>R | No |
ClinGen gnomAD |
|
|
rs1362425746 CA371853431 |
240 | T>A | No |
ClinGen gnomAD |
|
|
CA371853384 rs1269482156 |
246 | L>R | No |
ClinGen gnomAD |
|
|
CA4821785 rs746728067 |
247 | W>* | No |
ClinGen ExAC gnomAD |
|
|
rs745804050 CA4821782 |
249 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1338762257 CA371853369 |
249 | D>N | No |
ClinGen gnomAD |
|
|
CA371853361 rs1393163172 |
250 | D>N | No |
ClinGen Ensembl |
|
|
CA371853354 rs1485304076 |
251 | F>I | No |
ClinGen TOPMed |
|
|
rs1044970778 CA182774340 |
252 | T>I | No |
ClinGen TOPMed |
|
|
rs757271491 CA4821780 |
253 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1448180804 CA371853300 |
258 | C>F | No |
ClinGen gnomAD |
|
|
CA4821777 rs756524585 |
258 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs983939069 CA182774332 |
259 | L>F | No |
ClinGen Ensembl |
|
|
rs1245573773 CA371853294 |
259 | L>S | No |
ClinGen gnomAD |
|
|
CA182774330 rs763696181 |
260 | V>L | No |
ClinGen Ensembl |
|
|
CA371853278 rs1422915989 |
262 | N>H | No |
ClinGen TOPMed |
|
|
rs753043269 CA4821776 |
263 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs753043269 CA371853270 |
263 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1235335027 CA371853231 |
268 | T>I | No |
ClinGen gnomAD |
|
|
rs767919046 CA4821775 |
269 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1563784596 CA371853225 |
270 | T>A | No |
ClinGen Ensembl |
|
|
CA4821774 rs760017613 |
271 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA371853173 rs1300842216 |
275 | H>R | No |
ClinGen TOPMed |
|
|
CA371853175 rs1385825482 |
275 | H>Y | No |
ClinGen TOPMed |
|
|
CA4821750 rs763659968 |
276 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA371853168 rs547118456 |
276 | P>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4821751 rs547118456 |
276 | P>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs760305689 CA4821749 |
277 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs527380737 CA4821748 |
281 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs774294377 CA4821745 |
283 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs759288461 CA4821746 |
283 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759288461 CA371853122 |
283 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA182770624 rs1045946165 |
284 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1563760718 CA371853112 |
285 | S>P | No |
ClinGen Ensembl |
|
|
rs770905353 CA371853107 |
286 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821743 rs749218138 |
286 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs770905353 CA4821744 |
286 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777773209 CA4821742 |
288 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA4821741 rs374827150 |
288 | R>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs748322141 CA4821740 |
289 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA371853052 rs1185072443 |
294 | A>V | No |
ClinGen gnomAD |
|
|
CA371853050 rs1586912764 |
295 | M>V | No |
ClinGen Ensembl |
|
|
CA182770616 rs368118323 |
296 | E>D | No |
ClinGen ESP TOPMed |
|
|
CA4821737 rs747423317 |
301 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs1317615834 CA371852993 |
302 | H>Q | No |
ClinGen gnomAD |
|
|
rs1217960466 CA371852997 |
302 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs780530727 CA4821736 |
304 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA371852976 rs1174334333 |
305 | Q>* | No |
ClinGen TOPMed |
|
|
CA4821735 rs759044532 |
306 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA4821733 rs763527585 |
307 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755587989 CA4821732 |
307 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371852954 rs1346113264 |
308 | E>D | No |
ClinGen TOPMed |
|
|
CA371852943 rs1387321165 |
310 | E>A | No |
ClinGen gnomAD |
|
|
rs752325692 CA4821731 |
310 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA182770605 rs948590957 |
312 | E>D | No |
ClinGen TOPMed |
|
|
CA371852930 rs1470741154 |
312 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
COSM1102921 CA4821729 rs759167472 |
316 | S>L | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs767110089 CA4821730 |
316 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA4821710 rs368388279 |
317 | D>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA371851844 rs1246183655 |
318 | E>V | No |
ClinGen gnomAD |
|
|
CA371851819 rs1186674563 |
322 | D>H | No |
ClinGen gnomAD |
|
|
CA371851791 rs1462719916 |
326 | M>V | No |
ClinGen gnomAD |
|
|
CA371851760 rs1253429724 |
330 | S>N | No |
ClinGen gnomAD |
|
|
CA371851753 rs1586833258 |
331 | V>M | No |
ClinGen Ensembl |
|
|
CA182764462 rs974748998 |
332 | E>D | No |
ClinGen gnomAD |
|
|
CA371851729 rs1247462781 |
334 | V>G | No |
ClinGen gnomAD |
|
|
rs762767274 COSM270249 CA4821706 |
335 | G>D | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1393907753 CA371851720 |
336 | T>N | No |
ClinGen gnomAD |
|
|
CA4821703 rs374974168 |
337 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4821704 rs765265719 |
337 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs200907713 CA4821701 |
338 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4821702 rs776913859 |
338 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA371851705 rs1165415842 |
339 | A>S | No |
ClinGen gnomAD |
|
|
CA4821700 rs747251874 |
340 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs775790253 CA4821699 |
341 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs202028661 CA4821698 |
342 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA371851679 rs1456450036 |
343 | M>I | No |
ClinGen TOPMed |
|
|
rs771509402 CA4821695 |
343 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370561303 CA4821696 |
343 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA371851640 rs1372946741 |
349 | T>A | No |
ClinGen TOPMed gnomAD |
|
|
rs780693454 CA4821693 |
349 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1301013549 CA371851633 |
350 | M>T | No |
ClinGen TOPMed |
|
|
COSM1266933 CA182764461 rs908567268 |
350 | M>V | oesophagus [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA4821692 rs377506715 |
351 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs760363706 CA4821690 |
356 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371851588 rs1430855193 |
356 | T>S | No |
ClinGen gnomAD |
|
|
CA4821688 rs750268928 |
359 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1563727806 CA371851567 |
359 | E>K | No |
ClinGen Ensembl |
|
|
CA182764459 rs1037209658 |
360 | S>Y | No |
ClinGen TOPMed |
|
|
rs1408936576 CA371851555 |
361 | D>N | No |
ClinGen gnomAD |
|
|
rs1390002585 CA371851528 |
365 | M>T | No |
ClinGen gnomAD |
|
|
CA4821684 rs753775021 |
365 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs764276576 CA4821683 |
366 | V>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1365697292 CA371851494 |
370 | E>K | No |
ClinGen gnomAD |
|
|
CA371851484 rs1250035123 |
371 | D>G | No |
ClinGen gnomAD |
|
|
rs369783350 CA4821682 |
371 | D>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4821681 rs775878140 |
372 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371851467 rs1586832732 |
373 | E>V | No |
ClinGen Ensembl |
|
|
CA371851461 rs1289794117 |
374 | E>A | No |
ClinGen gnomAD |
|
|
CA371851450 rs759645713 |
375 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772387341 CA4821680 |
375 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371851446 rs771142783 |
376 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs560205328 CA4821678 |
376 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs771142783 CA4821677 |
376 | D>V | No |
ClinGen ExAC gnomAD |
|
|
CA371851443 rs1284949342 |
377 | G>R | No |
ClinGen gnomAD |
|
|
CA182764456 rs778211075 |
378 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821675 rs778211075 |
378 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821676 rs778211075 |
378 | T>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371851433 rs1330381946 |
379 | M>V | No |
ClinGen gnomAD |
|
|
CA371851423 rs1563727510 |
380 | K>E | No |
ClinGen Ensembl |
|
|
CA371851421 rs1321523112 |
380 | K>R | No |
ClinGen gnomAD |
|
|
rs1327011454 CA371852872 |
382 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs759861263 CA4821659 |
384 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA371852852 rs1563702897 |
385 | S>L | No |
ClinGen Ensembl |
|
|
CA4821658 rs142241647 |
386 | P>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs763253379 CA4821656 |
387 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371852843 rs763253379 |
387 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA371852829 rs1415359087 |
389 | Q>R | No |
ClinGen gnomAD |
|
|
rs1424860490 CA371852825 |
390 | R>G | No |
ClinGen gnomAD |
|
|
rs1176079716 CA371852823 |
390 | R>I | No |
ClinGen gnomAD |
|
|
rs1563702776 CA371852816 |
391 | P>S | No |
ClinGen Ensembl |
|
|
rs773542636 CA371852797 |
394 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821655 rs773542636 |
394 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1187315994 CA371852790 |
395 | D>N | No |
ClinGen gnomAD |
|
|
CA4821653 rs770322622 |
397 | F>V | No |
ClinGen ExAC gnomAD |
|
|
rs372444294 CA182760948 |
398 | D>G | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA371852759 rs1210425693 |
399 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA4821652 rs368988843 |
404 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1276226100 CA371852709 |
405 | K>R | No |
ClinGen gnomAD |
|
|
CA4821650 rs552761087 |
407 | H>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA371852684 rs1249444212 |
408 | E>D | No |
ClinGen TOPMed |
|
|
rs1190611897 CA371852687 |
408 | E>G | No |
ClinGen TOPMed |
|
|
COSM3835206 rs545643657 CA4821648 |
408 | E>K | breast [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA371852670 rs1449689641 |
410 | C>Y | No |
ClinGen TOPMed |
|
|
CA4821647 rs376001012 |
411 | N>S | No |
ClinGen ESP ExAC gnomAD |
|
|
CA4821646 rs749145486 |
412 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821645 rs577057842 |
413 | N>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs371715105 CA371852647 |
413 | N>K | No |
ClinGen ESP ExAC TOPMed |
|
|
CA4821642 rs369763923 |
414 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4821643 rs752670614 |
414 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA4821640 rs754970839 |
417 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
VAR_051670 rs36047674 CA4821639 |
418 | F>C | No |
ClinGen UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs536624746 CA4821637 |
420 | M>I | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs200371466 CA4821638 |
420 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA182760945 rs907796486 |
422 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs765569770 CA4821635 |
424 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs776939644 CA4821633 |
427 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821634 rs762318051 |
427 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA371852550 rs1273771278 |
428 | N>Y | No |
ClinGen gnomAD |
|
|
rs199671137 CA4821632 |
429 | W>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4821631 rs745324819 |
430 | K>T | No |
ClinGen ExAC gnomAD |
|
|
rs773893604 CA4821630 |
433 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA4821629 rs554676158 |
433 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs775921874 CA182760943 |
434 | D>G | No |
ClinGen gnomAD |
|
|
rs748944234 CA4821628 |
435 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1371596952 CA371852493 |
436 | D>E | No |
ClinGen gnomAD |
|
|
rs913664536 CA182744183 |
441 | K>E | No |
ClinGen TOPMed |
|
|
rs1481303387 CA371852215 |
444 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1165890095 CA371852186 |
447 | E>* | No |
ClinGen TOPMed |
|
|
CA4821610 rs770553429 |
451 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821611 rs770553429 |
451 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821612 rs761214264 |
451 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772958070 CA4821608 |
453 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1586601579 CA371852129 |
454 | A>S | No |
ClinGen Ensembl |
|
|
rs747942047 CA4821606 |
456 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA371852111 rs1586601543 |
457 | P>S | No |
ClinGen Ensembl |
|
|
CA371852105 rs1386682197 |
458 | M>K | No |
ClinGen gnomAD |
|
|
rs781008057 CA4821605 |
458 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746947660 CA4821603 COSM1102913 |
461 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA4821604 rs200670480 |
461 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1028037134 CA182744166 |
465 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA4821602 rs780118311 |
466 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA182744162 rs750630819 |
467 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs758402378 CA4821601 |
467 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA182744160 rs758402378 |
467 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA182744157 rs981667264 |
469 | R>K | No |
ClinGen Ensembl |
|
|
rs750678047 CA4821600 |
470 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs779061311 CA4821599 |
471 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4821598 rs757483119 |
472 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1421649694 CA371852011 COSM1458839 |
472 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA371851980 rs1465415994 |
477 | I>V | No |
ClinGen gnomAD |
|
|
rs1207020028 CA371851964 |
479 | D>V | No |
ClinGen gnomAD |
|
|
rs1484388193 CA371851960 |
480 | A>T | No |
ClinGen gnomAD |
|
|
CA4821595 COSM1102911 rs761145064 |
480 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA182744145 rs370627525 |
481 | M>T | No |
ClinGen ESP |
|
|
CA371851931 rs1408718916 |
484 | K>R | No |
ClinGen gnomAD |
|
|
CA371851930 rs1408718916 |
484 | K>T | No |
ClinGen gnomAD |
|
|
CA918324422 rs1586601195 |
492 | F>E | No |
ClinGen Ensembl |
|
|
CA371851868 rs1300464561 |
492 | F>S | No |
ClinGen gnomAD |
No associated diseases with Q13188
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| identical protein binding | Binding to an identical protein or proteins. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activator activity | Binds to and increases the activity of a protein serine/threonine kinase. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
26 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cell differentiation involved in embryonic placenta development | The process in which a relatively unspecialized cell acquires specialized features of the embryonic placenta. |
| central nervous system development | The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain and spinal cord. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord. |
| endocardium development | The process whose specific outcome is the progression of the endocardium over time, from its formation to the mature structure. The endocardium is an anatomical structure comprised of an endothelium and an extracellular matrix that forms the innermost layer of tissue of the heart, and lines the heart chambers. |
| hepatocyte apoptotic process | Any apoptotic process in a hepatocyte, the main structural component of the liver. |
| hippo signaling | The series of molecular signals mediated by the serine/threonine kinase Hippo or one of its orthologs. In Drosophila, Hippo in complex with the scaffold protein Salvador (Sav), phosphorylates and activates Warts (Wts), which in turn phosphorylates and inactivates the Yorkie (Yki) transcriptional activator. The core fly components hippo, sav, wts and mats are conserved in mammals as STK4/3 (MST1/2), SAV1/WW45, LATS1/2 and MOB1. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of canonical Wnt signaling pathway | Any process that decreases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of organ growth | Any process that stops, prevents, or reduces the frequency, rate or extent of growth of an organ of an organism. |
| neural tube formation | The formation of a tube from the flat layer of ectodermal cells known as the neural plate. This will give rise to the central nervous system. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of DNA-binding transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| positive regulation of extrinsic apoptotic signaling pathway via death domain receptors | Any process that activates or increases the frequency, rate or extent of extrinsic apoptotic signaling pathway via death domain receptors. |
| positive regulation of fat cell differentiation | Any process that activates or increases the frequency, rate or extent of adipocyte differentiation. |
| positive regulation of JNK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of protein kinase B signaling | Any process that activates or increases the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| primitive hemopoiesis | A first transient wave of blood cell production that, in vertebrates, gives rise to erythrocytes (red blood cells) and myeloid cells. |
| protein import into nucleus | The directed movement of a protein from the cytoplasm to the nucleus. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| protein stabilization | Any process involved in maintaining the structure and integrity of a protein and preventing it from degradation or aggregation. |
| protein tetramerization | The formation of a protein tetramer, a macromolecular structure consisting of four noncovalently associated identical or nonidentical subunits. |
| regulation of cell differentiation involved in embryonic placenta development | Any process that modulates the rate, frequency or extent of cell differentiation that contributes to the progression of the placenta over time, from its initial condition to its mature state. |
| regulation of MAPK cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAP kinase (MAPK) cascade. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
24 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9L6 | STK4 | Serine/threonine-protein kinase 4 | Bos taurus (Bovine) | SS |
| Q3SWY6 | STK25 | Serine/threonine-protein kinase 25 | Bos taurus (Bovine) | PR |
| Q5ZJK4 | STK4 | Serine/threonine-protein kinase 4 | Gallus gallus (Chicken) | SS |
| Q13043 | STK4 | Serine/threonine-protein kinase 4 | Homo sapiens (Human) | EV |
| Q9NQU5 | PAK6 | Serine/threonine-protein kinase PAK 6 | Homo sapiens (Human) | EV |
| Q9P286 | PAK5 | Serine/threonine-protein kinase PAK 5 | Homo sapiens (Human) | EV |
| O96013 | PAK4 | Serine/threonine-protein kinase PAK 4 | Homo sapiens (Human) | EV |
| Q13177 | PAK2 | Serine/threonine-protein kinase PAK 2 | Homo sapiens (Human) | EV |
| O75914 | PAK3 | Serine/threonine-protein kinase PAK 3 | Homo sapiens (Human) | SS |
| Q13153 | PAK1 | Serine/threonine-protein kinase PAK 1 | Homo sapiens (Human) | EV |
| O95819 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | Homo sapiens (Human) | PR |
| Q9Y6E0 | STK24 | Serine/threonine-protein kinase 24 | Homo sapiens (Human) | PR |
| O00506 | STK25 | Serine/threonine-protein kinase 25 | Homo sapiens (Human) | PR |
| Q9JI11 | Stk4 | Serine/threonine-protein kinase 4 | Mus musculus (Mouse) | PR |
| Q9JI10 | Stk3 | Serine/threonine-protein kinase 3 | Mus musculus (Mouse) | SS |
| Q9Z2W1 | Stk25 | Serine/threonine-protein kinase 25 | Mus musculus (Mouse) | PR |
| Q99JT2 | Stk26 | Serine/threonine-protein kinase 26 | Mus musculus (Mouse) | PR |
| Q99KH8 | Stk24 | Serine/threonine-protein kinase 24 | Mus musculus (Mouse) | PR |
| O54748 | Stk3 | Serine/threonine-protein kinase 3 | Rattus norvegicus (Rat) | SS |
| A4K2T0 | STK4 | Serine/threonine-protein kinase 4 | Macaca mulatta (Rhesus macaque) | PR |
| H2L099 | gck-1 | Germinal center kinase 1 | Caenorhabditis elegans | PR |
| Q9NB31 | cst-1 | Serine/threonine-protein kinase cst-1 | Caenorhabditis elegans | PR |
| Q6P3Q4 | stk4 | Serine/threonine-protein kinase 4 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7ZUQ3 | stk3 | Serine/threonine-protein kinase 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEQPPAPKSK | LKKLSEDSLT | KQPEEVFDVL | EKLGEGSYGS | VFKAIHKESG | QVVAIKQVPV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ESDLQEIIKE | ISIMQQCDSP | YVVKYYGSYF | KNTDLWIVME | YCGAGSVSDI | IRLRNKTLIE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DEIATILKST | LKGLEYLHFM | RKIHRDIKAG | NILLNTEGHA | KLADFGVAGQ | LTDTMAKRNT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VIGTPFWMAP | EVIQEIGYNC | VADIWSLGIT | SIEMAEGKPP | YADIHPMRAI | FMIPTNPPPT |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FRKPELWSDD | FTDFVKKCLV | KNPEQRATAT | QLLQHPFIKN | AKPVSILRDL | ITEAMEIKAK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| RHEEQQRELE | EEEENSDEDE | LDSHTMVKTS | VESVGTMRAT | STMSEGAQTM | IEHNSTMLES |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DLGTMVINSE | DEEEEDGTMK | RNATSPQVQR | PSFMDYFDKQ | DFKNKSHENC | NQNMHEPFPM |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SKNVFPDNWK | VPQDGDFDFL | KNLSLEELQM | RLKALDPMME | REIEELRQRY | TAKRQPILDA |
| 490 | |||||
| MDAKKRRQQN | F |