Q0VC18
Gene name |
ARL4D |
Protein name |
ADP-ribosylation factor-like protein 4D |
Names |
|
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:538936 |
EC number |
|
Protein Class |
|
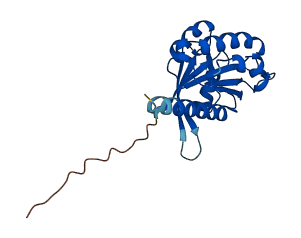
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q0VC18
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q0VC18-F1 | Predicted | AlphaFoldDB |
72 variants for Q0VC18
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs448931729 | 14 | F>C | No | EVA | |
| rs433885601 | 34 | T>A | No | EVA | |
| rs453993553 | 41 | K>* | No | EVA | |
| rs455870914 | 57 | E>D | No | EVA | |
| rs475907241 | 61 | V>G | No | EVA | |
| rs444479917 | 63 | L>P | No | EVA | |
| rs458284506 | 64 | G>W | No | EVA | |
| rs478417888 | 70 | T>N | No | EVA | |
| rs440688442 | 73 | V>M | No | EVA | |
| rs480058762 | 79 | Q>H | No | EVA | |
| rs448792991 | 82 | L>I | No | EVA | |
| rs482795060 | 86 | W>L | No | EVA | |
| rs462502483 | 86 | W>R | No | EVA | |
| rs444892271 | 87 | R>G | No | EVA | |
| rs447620450 | 96 | L>R | No | EVA | |
| rs433737930 | 96 | L>V | No | EVA | |
| rs435629151 | 97 | V>E | No | EVA | |
| rs435629151 | 97 | V>G | No | EVA | |
| rs467661962 | 97 | V>M | No | EVA | |
| rs455734355 | 98 | F>I | No | EVA | |
| rs475810953 | 98 | F>L | No | EVA | |
| rs438102191 | 101 | D>A | No | EVA | |
| rs451752474 | 102 | A>S | No | EVA | |
| rs472004347 | 102 | A>V | No | EVA | |
| rs460728078 | 105 | A>V | No | EVA | |
| rs474312512 | 106 | E>Q | No | EVA | |
| rs443041716 | 107 | R>P | No | EVA | |
| rs482641273 | 108 | L>M | No | EVA | |
| rs444919462 | 109 | E>V | No | EVA | |
| rs458631559 | 110 | E>G | No | EVA | |
| rs458631559 | 110 | E>V | No | EVA | |
| rs447481633 | 111 | A>G | No | EVA | |
| rs478849597 | 111 | A>P | No | EVA | |
| rs478849597 | 111 | A>S | No | EVA | |
| rs436289474 | 112 | K>R | No | EVA | |
| rs449933790 | 113 | V>G | No | EVA | |
| rs437970576 | 114 | E>G | No | EVA | |
| rs469379762 | 114 | E>K | No | EVA | |
| rs451747920 | 115 | L>F | No | EVA | |
| rs471703938 | 115 | L>P | No | EVA | |
| rs451747920 | 115 | L>V | No | EVA | |
| rs434006269 | 116 | H>P | No | EVA | |
| rs454186729 | 117 | R>G | No | EVA | |
| rs474169652 | 117 | R>Q | No | EVA | |
| rs443102732 | 118 | I>T | No | EVA | |
| rs456796859 | 119 | S>R | No | EVA | |
| rs438377371 | 121 | A>S | No | EVA | |
| rs458628014 | 123 | D>A | No | EVA | |
| rs440904579 | 129 | V>G | No | EVA | |
| rs461160899 | 131 | V>G | No | EVA | |
| rs481185915 | 133 | A>P | No | EVA | |
| rs449932496 | 145 | A>P | No | EVA | |
| rs449932496 | 145 | A>S | No | EVA | |
| rs470083534 | 146 | E>G | No | EVA | |
| rs483138829 | 147 | V>G | No | EVA | |
| rs465158008 | 152 | A>G | No | EVA | |
| rs135592535 | 153 | V>G | No | EVA | |
| rs454047841 | 163 | H>P | No | EVA | |
| rs467832166 | 164 | V>A | No | EVA | |
| rs456816904 | 169 | A>V | No | EVA | |
| rs876408996 | 170 | V>G | No | EVA | |
| rs876463155 | 172 | G>R | No | EVA | |
| rs476937670 | 173 | L>P | No | EVA | |
| rs434785160 | 181 | R>H | No | EVA | |
| rs439038351 | 183 | Y>D | No | EVA | |
| rs452092671 | 184 | E>G | No | EVA | |
| rs440965839 | 196 | G>A | No | EVA | |
| rs472156038 | 196 | G>R | No | EVA | |
| rs461022083 | 198 | K>R | No | EVA | |
| rs481249418 | 199 | R>G | No | EVA | |
| rs463471436 | 201 | R>C | No | EVA | |
| rs443405787 | 201 | R>R | No | EVA |
No associated diseases with Q0VC18
5 regional properties for Q0VC18
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 18 - 294 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 23 - 287 | IPR001245 |
| domain | CARD domain | 435 - 513 | IPR001315 |
| active_site | Serine/threonine-protein kinase, active site | 142 - 154 | IPR008271 |
| domain | RIP2, CARD domain | 437 - 522 | IPR042149 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
47 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q5E9I6 | ARF3 | ADP-ribosylation factor 3 | Bos taurus (Bovine) | SS |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGNHLTEMAP | TTSFLPHFQA | LHVVVIGLDS | AGKTSLLYRL | KFKEFVQSIP | TKGFNTEKIR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VPLGGSRGIT | FQVWDVGGQE | KLRPLWRSYT | RRTDGLVFVV | DAAEAERLEE | AKVELHRISR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ASDNQGVPVL | VLANKQDQPG | ALSAAEVEKR | LAVRELATAT | LTHVQGCSAV | DGLGLQPGLE |
| 190 | |||||
| RLYEMILKRK | KAARAGKKRR |