Q0JC44
Gene name |
CML22 (Os04g0492800, LOC_Os04g41540, OJ990528_30.6, OSJNBb0091E11.1) |
Protein name |
Probable calcium-binding protein CML22 |
Names |
Calmodulin-like protein 22 |
Species |
Oryza sativa subsp japonica (Rice) |
KEGG Pathway |
osa:4336258 |
EC number |
|
Protein Class |
|
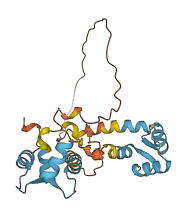
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q0JC44
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q0JC44-F1 | Predicted | AlphaFoldDB |
No variants for Q0JC44
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for Q0JC44 | |||||
No associated diseases with Q0JC44
6 regional properties for Q0JC44
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | EF-hand domain | 88 - 159 | IPR002048-1 |
| domain | EF-hand domain | 174 - 247 | IPR002048-2 |
| binding_site | EF-Hand 1, calcium-binding site | 101 - 113 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 137 - 149 | IPR018247-2 |
| binding_site | EF-Hand 1, calcium-binding site | 187 - 199 | IPR018247-3 |
| binding_site | EF-Hand 1, calcium-binding site | 225 - 237 | IPR018247-4 |
No GO annotations of cellular component
| Name | Definition |
|---|---|
| No GO annotations for cellular component |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
5 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q80X60 | Efcab3 | EF-hand calcium-binding domain-containing protein 3 | Mus musculus (Mouse) | PR |
| Q6P1E8 | Efcab6 | EF-hand calcium-binding domain-containing protein 6 | Mus musculus (Mouse) | PR |
| Q66HC0 | Efcab3 | EF-hand calcium-binding domain-containing protein 3 | Rattus norvegicus (Rat) | PR |
| Q9FYK2 | CML25 | Probable calcium-binding protein CML25 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRN6 | CML22 | Probable calcium-binding protein CML22 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSCQVPTVPA | SPVVKFPLLP | RGLLSYLPAN | LSSILPVARG | AASTCEASST | TTTTMPPEPP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ASPPPPKKMS | PPGAGAGAGS | KKKQQQQADA | AELARVFELF | DRNGDGRITR | EELEDSLGKL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GIPVPADELA | AVIARIDANG | DGCVDVEEFG | ELYRSIMAGG | DDSKDGRAKE | EEEEEDGDMR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EAFRVFDANG | DGYITVDELG | AVLASLGLKQ | GRTAEECRRM | IGQVDRDGDG | RVDFHEFLQM |
| MRGGGFAALG |