Q08E52
Gene name |
PAK1 |
Protein name |
Serine/threonine-protein kinase PAK 1 |
Names |
Alpha-PAK, p21-activated kinase 1, PAK-1, p65-PAK |
Species |
Bos taurus (Bovine) |
KEGG Pathway |
bta:533729 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
269-520 (Protein kinase domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
405-428 (Activation loop from InterPro)
Target domain |
269-520 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Lei M et al. (2000) "Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch", Cell, 102, 387-97
- Totaro A et al. (2007) "Identification of an intramolecular interaction important for the regulation of GIT1 functions", Molecular biology of the cell, 18, 5124-38
- Bautista L et al. (2020) "p21-Activated Kinases in Thyroid Cancer", Endocrinology, 161,
- Chong C et al. (2001) "The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity", The Journal of biological chemistry, 276, 17347-53
- Wang J et al. (2011) "Structural insights into the autoactivation mechanism of p21-activated protein kinase", Structure (London, England : 1993), 19, 1752-61
- Ha BH et al. (2012) "Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate", Proceedings of the National Academy of Sciences of the United States of America, 109, 16107-12
- Rousseau V et al. (2003) "A new constitutively active brain PAK3 isoform displays modified specificities toward Rac and Cdc42 GTPases", The Journal of biological chemistry, 278, 3912-20
- Ching YP et al. (2003) "Identification of an autoinhibitory domain of p21-activated protein kinase 5", The Journal of biological chemistry, 278, 33621-4
- Dan C et al. (2002) "PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells", Molecular and cellular biology, 22, 567-77
- Kaur R et al. (2005) "Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase", The Journal of biological chemistry, 280, 3323-30
- Gao J et al. (2013) "Substrate and inhibitor specificity of the type II p21-activated kinase, PAK6", PloS one, 8, e77818
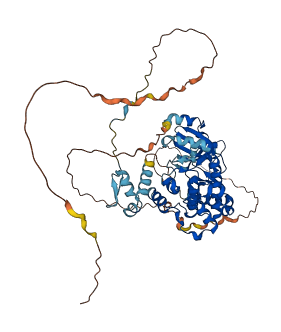
Autoinhibited structure

Activated structure

1 structures for Q08E52
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q08E52-F1 | Predicted | AlphaFoldDB |
50 variants for Q08E52
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs464663376 | 2 | S>P | No | Ensembl | |
| rs434784910 | 8 | I>L | No | Ensembl | |
| rs446780180 | 10 | D>V | No | Ensembl | |
| rs468555032 | 14 | A>S | No | Ensembl | |
| rs435675974 | 19 | N>T | No | Ensembl | |
| rs476449025 | 33 | T>N | No | Ensembl | |
| rs436951522 | 34 | L>P | No | Ensembl | |
| rs470867353 | 38 | S>T | No | Ensembl | |
| rs440869923 | 40 | P>S | No | Ensembl | |
| rs474739659 | 55 | Y>F | No | Ensembl | |
| rs441713823 | 58 | I>F | No | Ensembl | |
| rs463367017 | 58 | I>N | No | Ensembl | |
| rs458306449 | 59 | L>F | No | Ensembl | |
| rs479917392 | 61 | G>E | No | Ensembl | |
| rs435534083 | 62 | D>E | No | Ensembl | |
| rs446843277 | 62 | D>N | No | Ensembl | |
| rs468418229 | 62 | D>V | No | Ensembl | |
| rs470540309 | 100 | P>Q | No | Ensembl | |
| rs440945273 | 111 | N>T | No | Ensembl | |
| rs459351052 | 135 | K>T | No | Ensembl | |
| rs481681895 | 140 | Q>* | No | Ensembl | |
| rs444184903 | 160 | N>D | No | Ensembl | |
| rs462710534 | 160 | N>T | No | Ensembl | |
| rs800484308 | 171 | P>S | No | Ensembl | |
| rs471506498 | 185 | P>Q | No | Ensembl | |
| rs446120180 | 233 | A>S | No | Ensembl | |
| rs381763110 | 236 | R>W | No | Ensembl | |
| rs448524116 | 279 | A>V | No | Ensembl | |
| rs470085042 | 282 | T>P | No | Ensembl | |
| rs465126576 | 283 | V>A | No | Ensembl | |
| rs456256881 | 283 | V>M | No | Ensembl | |
| rs432274731 | 284 | Y>* | No | Ensembl | |
| rs453970222 | 285 | T>P | No | Ensembl | |
| rs472203003 | 286 | A>S | No | Ensembl | |
| rs435954308 | 288 | D>E | No | Ensembl | |
| rs454671896 | 289 | V>L | No | Ensembl | |
| rs443274900 | 290 | A>P | No | Ensembl | |
| rs459086764 | 291 | T>P | No | Ensembl | |
| rs441510070 | 293 | Q>K | No | Ensembl | |
| rs459859364 | 293 | Q>R | No | Ensembl | |
| rs448386707 | 294 | E>G | No | Ensembl | |
| rs481246670 | 294 | E>Q | No | Ensembl | |
| rs460217536 | 327 | V>L | No | Ensembl | |
| rs518227183 | 367 | A>S | No | Ensembl | |
| rs458690512 | 520 | L>R | No | Ensembl | |
| rs477320047 | 524 | K>M | No | Ensembl | |
| rs447454650 | 524 | K>N | No | Ensembl | |
| rs459382671 | 525 | P>S | No | Ensembl | |
| rs478535217 | 531 | P>L | No | Ensembl | |
| rs481057682 | 542 | N>S | No | Ensembl |
No associated diseases with Q08E52
4 regional properties for Q08E52
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin filament | A filamentous structure formed of a two-stranded helical polymer of the protein actin and associated proteins. Actin filaments are a major component of the contractile apparatus of skeletal muscle and the microfilaments of the cytoskeleton of eukaryotic cells. The filaments, comprising polymerized globular actin molecules, appear as flexible structures with a diameter of 5-9 nm. They are organized into a variety of linear bundles, two-dimensional networks, and three dimensional gels. In the cytoskeleton they are most highly concentrated in the cortex of the cell just beneath the plasma membrane. |
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| chromosome | A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| microtubule organizing center | An intracellular structure that can catalyze gamma-tubulin-dependent microtubule nucleation and that can anchor microtubules by interacting with their minus ends, plus ends or sides. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| ruffle membrane | The portion of the plasma membrane surrounding a ruffle. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| collagen binding | Binding to collagen, a group of fibrous proteins of very high tensile strength that form the main component of connective tissue in animals. Collagen is highly enriched in glycine (some regions are 33% glycine) and proline, occurring predominantly as 3-hydroxyproline (about 20%). |
| gamma-tubulin binding | Binding to the microtubule constituent protein gamma-tubulin. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| small GTPase binding | Binding to a small monomeric GTPase. |
25 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton reorganization | A process that is carried out at the cellular level which results in dynamic structural changes to the arrangement of constituent parts of cytoskeletal structures comprising actin filaments and their associated proteins. |
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| branching morphogenesis of an epithelial tube | The process in which the anatomical structures of branches in an epithelial tube are generated and organized. A tube is a long hollow cylinder. |
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| cellular response to DNA damage stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| chromatin remodeling | A dynamic process of chromatin reorganization resulting in changes to chromatin structure. These changes allow DNA metabolic processes such as transcriptional regulation, DNA recombination, DNA repair, and DNA replication. |
| exocytosis | A process of secretion by a cell that results in the release of intracellular molecules (e.g. hormones, matrix proteins) contained within a membrane-bounded vesicle. Exocytosis can occur either by full fusion, when the vesicle collapses into the plasma membrane, or by a kiss-and-run mechanism that involves the formation of a transient contact, a pore, between a granule (for exemple of chromaffin cells) and the plasma membrane. The latter process most of the time leads to only partial secretion of the granule content. Exocytosis begins with steps that prepare vesicles for fusion with the membrane (tethering and docking) and ends when molecules are secreted from the cell. |
| hepatocyte growth factor receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a hepatocyte growth factor receptor, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| negative regulation of cell proliferation involved in contact inhibition | Any process that stops, prevents or reduces the rate or extent of cell proliferation in response to cell density. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of intracellular estrogen receptor signaling pathway | Any process that activates or increases the frequency, rate or extent of the activity of an intracellular estrogen receptor signaling pathway. |
| positive regulation of JUN kinase activity | Any process that activates or increases the frequency, rate or extent of JUN kinase activity. |
| positive regulation of microtubule nucleation | Any process that increases the rate, frequency or extent of microtubule nucleation. Microtubule nucleation is the 'de novo' formation of a microtubule, in which tubulin heterodimers form metastable oligomeric aggregates, some of which go on to support formation of a complete microtubule. Microtubule nucleation usually occurs from a specific site within a cell. |
| positive regulation of peptidyl-serine phosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation of peptidyl-serine. |
| positive regulation of protein phosphorylation | Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to amino acids within a protein. |
| positive regulation of stress fiber assembly | Any process that activates or increases the frequency, rate or extent of the assembly of a stress fiber, a bundle of microfilaments and other proteins found in fibroblasts. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of actin cytoskeleton organization | Any process that modulates the frequency, rate or extent of the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| regulation of axonogenesis | Any process that modulates the frequency, rate or extent of axonogenesis, the generation of an axon, the long process of a neuron. |
| regulation of MAPK cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAP kinase (MAPK) cascade. |
| wound healing | The series of events that restore integrity to a damaged tissue, following an injury. |
22 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q12469 | SKM1 | Serine/threonine-protein kinase SKM1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q5E9L6 | STK4 | Serine/threonine-protein kinase 4 | Bos taurus (Bovine) | SS |
| Q7YQL4 | PAK3 | Serine/threonine-protein kinase PAK 3 | Pan troglodytes (Chimpanzee) | SS |
| Q9VXE5 | mbt | Serine/threonine-protein kinase PAK mbt | Drosophila melanogaster (Fruit fly) | PR |
| Q9P286 | PAK5 | Serine/threonine-protein kinase PAK 5 | Homo sapiens (Human) | EV |
| Q9NQU5 | PAK6 | Serine/threonine-protein kinase PAK 6 | Homo sapiens (Human) | EV |
| O96013 | PAK4 | Serine/threonine-protein kinase PAK 4 | Homo sapiens (Human) | EV |
| O75914 | PAK3 | Serine/threonine-protein kinase PAK 3 | Homo sapiens (Human) | SS |
| Q13177 | PAK2 | Serine/threonine-protein kinase PAK 2 | Homo sapiens (Human) | EV |
| Q13153 | PAK1 | Serine/threonine-protein kinase PAK 1 | Homo sapiens (Human) | EV |
| Q8C015 | Pak5 | Serine/threonine-protein kinase PAK 5 | Mus musculus (Mouse) | SS |
| Q3ULB5 | Pak6 | Serine/threonine-protein kinase PAK 6 | Mus musculus (Mouse) | PR |
| Q61036 | Pak3 | Serine/threonine-protein kinase PAK 3 | Mus musculus (Mouse) | SS |
| Q8CIN4 | Pak2 | Serine/threonine-protein kinase PAK 2 | Mus musculus (Mouse) | SS |
| O88643 | Pak1 | Serine/threonine-protein kinase PAK 1 | Mus musculus (Mouse) | SS |
| Q62829 | Pak3 | Serine/threonine-protein kinase PAK 3 | Rattus norvegicus (Rat) | SS |
| D4A280 | Pak5 | Serine/threonine-protein kinase PAK 5 | Rattus norvegicus (Rat) | SS |
| Q64303 | Pak2 | Serine/threonine-protein kinase PAK 2 | Rattus norvegicus (Rat) | SS |
| P35465 | Pak1 | Serine/threonine-protein kinase PAK 1 | Rattus norvegicus (Rat) | SS |
| G5EFU0 | pak-2 | Serine/threonine-protein kinase pak-2 | Caenorhabditis elegans | PR |
| Q17850 | pak-1 | Serine/threonine-protein kinase pak-1 | Caenorhabditis elegans | PR |
| G5EGQ3 | max-2 | Serine/threonine-protein kinase max-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSNNGLDIQD | KPPAPPMRNT | STMIGAGSKD | AGTLNHGSKP | LPPNPEEKKK | KDRFYRAILP |
| 70 | 80 | 90 | 100 | 110 | 120 |
| GDKTNKKKEK | ERPEISLPSD | FEHTIHVGFD | AVTGEFTGMP | EQWARLLQTS | NITKSEQKKN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PQAVLDVLEF | YNSKKTSNSQ | KYMSFTDKSA | EDYNSSNTLN | VKAVSETPAV | PPVSEDEDDD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DDGTPPPVIA | PRPEHTKSVY | TRSVIEPLPI | TPTRDVATSP | ISPTENNTTP | PDALTRNTEK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QKKKPKMSDE | EILEKLRSIV | SVGDPKKKYT | RFEKIGQGAS | GTVYTAMDVA | TGQEVAIKQM |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NLQQQPKKEL | IINEILVMRE | NKNPNIVNYL | DSYLVGDELW | VVMEYLAGGS | LTDVVTETCM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DEGQIAAVCR | ECLQALEFLH | SNQVIHRDIK | SDNILLGMDG | SVKLTDFGFC | AQITPEQSKR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| STMVGTPYWM | APEVVTRKAY | GPKVDIWSLG | IMAIEMIEGE | PPYLNENPLR | ALYLIATNGT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PELQNPEKLS | AIFRDFLNRC | LEMDVEKRGS | AKELLQHQFL | KIAKPLSSLT | PLIAAAKEAT |
| KNNH |