Q08AT1
Gene name |
Rasl12 |
Protein name |
Ras-like protein family member 12 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:70784 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
|
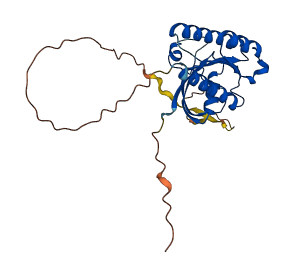
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q08AT1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q08AT1-F1 | Predicted | AlphaFoldDB |
12 variants for Q08AT1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389042182 | 27 | G>A | No | EVA | |
| rs3399230519 | 36 | L>Q | No | EVA | |
| rs3389033049 | 70 | L>Q | No | EVA | |
| rs3389000857 | 72 | V>L | No | EVA | |
| rs3389061625 | 144 | V>A | No | EVA | |
| rs3389055898 | 144 | V>I | No | EVA | |
| rs3389028841 | 151 | A>G | No | EVA | |
| rs3389028876 | 152 | L>F | No | EVA | |
| rs3389042174 | 156 | F>L | No | EVA | |
| rs3389054954 | 199 | S>T | No | EVA | |
| rs3389055907 | 207 | Q>P | No | EVA | |
| rs3389047798 | 251 | K>* | No | EVA |
No associated diseases with Q08AT1
1 regional properties for Q08AT1
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 21 - 165 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| G protein activity | A molecular function regulator that cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular processes. Intrinsic GTPase activity returns the G protein to its GDP-bound state. The return to the GDP-bound state can be accelerated by the action of a GTPase-activating protein (GAP). |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSSVFGKPRA | GSGPHSVPLE | VNLAILGRRG | AGKSALTVKF | LTKRFISEYD | PNLEDTYSSE |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ETVDHQPVHL | RVMDTADLDT | PRNCERYLNW | AHAFLVVYSV | DSRASFEGSS | SYLELLALHA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KETQRGYPAL | LLGNKLDMAQ | YRQVTKAEGA | ALAGRFGCLF | FEVSACLDFE | HVQHVFHEAV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| REVRRELDKS | PLARPLFISE | EKTLSHQTPL | TARHGLASCT | FNTLSTASLK | EMPTVAQAKL |
| 250 | 260 | ||||
| VTVKSSRAQS | KRKAPTLTLL | KGFKIF |