Q08209
Gene name |
PPP3CA |
Protein name |
Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform |
Names |
CAM-PRP catalytic subunit, Calmodulin-dependent calcineurin A subunit alpha isoform |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5530 |
EC number |
3.1.3.16: Phosphoric monoester hydrolases |
Protein Class |
SERINE/THREONINE-PROTEIN PHOSPHATASE 2B CATALYTIC SUBUNIT 1-RELATED (PTHR45673) |
Descriptions
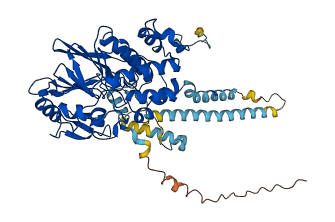
Calmodulin-stimulated protein phosphatase (Calcineurin in short) plays a central role in the transduction of intracellular Ca2+-mediated signals in processes involving T-cell activation, development and function of the central nervous system, and cardiac growth. Calcineurin is composed of a catalytic domain at the N terminus, B chain, a regulatory domain (RD), autoinhibitory domain (AID), and C-terminal domain (ATC). The catalytic domain is autoinhibited by the AID region in the absence of calcium and CaM. The conformation of autoinhibited Calcineurin is released by the binding of CaM to the RD with the presence of calcium, leading to activation of the phosphatase.
Autoinhibitory domains (AIDs)
Target domain |
1-348 (Calcineurin-like phosphoesterase domain) |
Relief mechanism |
Partner binding |
Assay |
Structural analysis |
Target domain |
1-348 (Calcineurin-like phosphoesterase domain) |
Relief mechanism |
Partner binding |
Assay |
Structural analysis |
Accessory elements
No accessory elements
References
- Rumi-Masante J et al. (2012) "Structural basis for activation of calcineurin by calmodulin", Journal of molecular biology, 415, 307-17
- Li SJ et al. (2016) "Cooperative autoinhibition and multi-level activation mechanisms of calcineurin", Cell research, 26, 336-49
- Kissinger CR et al. (1995) "Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex", Nature, 378, 641-4
- Ye Q et al. (2008) "The complex structure of calmodulin bound to a calcineurin peptide", Proteins, 73, 19-27
Autoinhibited structure

Activated structure

18 structures for Q08209
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1AUI | X-ray | 210 A | A | 1-521 | PDB |
| 1M63 | X-ray | 280 A | A/E | 1-372 | PDB |
| 1MF8 | X-ray | 310 A | A | 20-392 | PDB |
| 2JOG | NMR | - | A | 21-347 | PDB |
| 2JZI | NMR | - | B | 391-414 | PDB |
| 2P6B | X-ray | 230 A | A/C | 1-380 | PDB |
| 2R28 | X-ray | 186 A | C/D | 389-413 | PDB |
| 2W73 | X-ray | 145 A | K/L/M/O | 395-411 | PDB |
| 3LL8 | X-ray | 200 A | A/C | 14-370 | PDB |
| 4F0Z | X-ray | 170 A | A | 1-370 | PDB |
| 4Q5U | X-ray | 195 A | C | 391-414 | PDB |
| 5C1V | X-ray | 335 A | A/B | 2-346 | PDB |
| 5SVE | X-ray | 260 A | A | 1-370 | PDB |
| 6NUC | X-ray | 190 A | A | 1-370 | PDB |
| 6NUF | X-ray | 190 A | A | 1-370 | PDB |
| 6NUU | X-ray | 230 A | A | 1-370 | PDB |
| 6UUQ | X-ray | 185 A | A | 26-339 | PDB |
| AF-Q08209-F1 | Predicted | AlphaFoldDB |
206 variants for Q08209
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001027515 RCV001267106 CA357950659 rs1553925558 RCV000509875 VAR_080348 |
92 | H>R | Intellectual disability IECEE1 Epileptic encephalopathy, infantile or early childhood, 1 Inborn genetic diseases [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
VAR_080349 CA357948414 rs199706529 RCV000509768 |
281 | H>Q | IECEE1 Epileptic encephalopathy, infantile or early childhood, 1 [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ExAC dbSNP gnomAD |
|
RCV000624048 RCV000509989 VAR_080350 rs1553923787 CA357948412 |
282 | E>K | IECEE1 Epileptic encephalopathy, infantile or early childhood, 1 Inborn genetic diseases [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1553920383 RCV000677432 |
419 | S>missing | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1727004803 RCV001198048 |
429 | T>missing | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000735832 rs1560570541 |
431 | M>missing | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000625968 rs1553920379 |
438 | S>missing | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1578388765 RCV000987459 |
438 | S>missing | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA357947963 RCV000509789 rs1553920376 |
445 | Q>* | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001865669 RCV000510091 VAR_080352 rs1553920374 CA357947948 |
447 | A>T | Epileptic encephalopathy, infantile or early childhood, 1 IECEE1; unknown pathological significance [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
VAR_081902 CA357947765 RCV000735833 rs1560567347 |
470 | F>L | Arthrogryposis, cleft palate, craniosynostosis, and impaired intellectual development ACCIID [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA357947743 rs1560567337 RCV000735834 VAR_081903 |
473 | A>T | Arthrogryposis, cleft palate, craniosynostosis, and impaired intellectual development ACCIID [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001031007 rs1726628642 |
493 | D>N | Epileptic encephalopathy, infantile or early childhood, 1 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1175599344 CA357949688 |
2 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
CA357949683 rs1175599344 |
2 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs376661307 CA3024650 |
3 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs774503546 COSM1309570 CA3024651 |
3 | E>K | urinary_tract [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs749627735 CA3024649 |
4 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA3024647 rs770314812 |
6 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1730017441 RCV001090878 |
8 | D>Y | No |
ClinVar dbSNP |
|
|
rs777239762 CA3024645 |
9 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs746412506 CA3024646 |
9 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357949500 rs1384222183 |
13 | T>M | No |
ClinGen TOPMed |
|
|
CA357949512 rs1560730716 |
13 | T>P | No |
ClinGen Ensembl |
|
|
rs1232030786 CA357949495 |
14 | T>A | No |
ClinGen gnomAD |
|
|
rs1309941061 CA357949489 |
14 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA103004284 rs866171704 |
15 | D>N | No |
ClinGen Ensembl |
|
|
CA357949427 rs1219287075 |
18 | V>L | No |
ClinGen gnomAD |
|
|
CA357951132 rs1418346739 |
23 | F>I | No |
ClinGen gnomAD |
|
|
rs776490146 CA102987409 |
27 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357951102 rs1435984083 |
27 | H>Y | No |
ClinGen gnomAD |
|
|
CA3024564 rs746829234 |
28 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024565 rs770562478 |
28 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1724781554 RCV001312044 |
30 | T>I | No |
ClinVar dbSNP |
|
|
rs200056844 CA3024562 |
31 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs748137556 CA3024561 |
33 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs755119794 CA3024560 COSM447171 |
34 | V>A | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA3024559 rs755119794 |
34 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1054602875 CA102987408 |
34 | V>L | No |
ClinGen Ensembl |
|
|
CA357951051 rs1351508365 |
36 | D>N | No |
ClinGen gnomAD |
|
|
CA3024558 rs146174889 |
37 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA357951033 rs1457318131 |
38 | D>G | No |
ClinGen TOPMed |
|
|
rs780337368 CA3024557 |
38 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs996368040 CA102987407 |
39 | G>R | No |
ClinGen Ensembl |
|
|
CA3024556 rs756386250 |
41 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767812589 CA3024554 |
42 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs762386324 CA3024553 |
42 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762386324 CA357951008 |
42 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1160406789 CA357950987 |
45 | I>M | No |
ClinGen gnomAD |
|
|
rs1411992353 CA357950969 |
48 | A>S | No |
ClinGen gnomAD |
|
|
rs764832679 CA3024551 |
48 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA102987405 rs771036944 |
50 | L>F | No |
ClinGen Ensembl |
|
|
CA357950950 rs1258430839 |
51 | M>R | No |
ClinGen gnomAD |
|
|
CA357950937 rs1415693404 |
52 | K>N | No |
ClinGen TOPMed |
|
|
CA357950891 rs1191653592 |
59 | S>N | No |
ClinGen gnomAD |
|
|
rs1191653592 CA357950890 |
59 | S>T | No |
ClinGen gnomAD |
|
|
CA102987404 rs368190241 |
61 | A>T | No |
ClinGen ESP |
|
|
CA3024547 rs760289268 |
64 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA357950844 rs1243795414 |
66 | T>R | No |
ClinGen gnomAD |
|
|
rs145968865 CA3024545 |
71 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs141696639 CA3024543 |
73 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
COSM199991 CA3024544 rs141696639 |
73 | R>Q | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA357950801 rs768666798 |
74 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024542 rs768666798 |
74 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024541 rs749393129 |
74 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
CA3024540 rs371141456 |
76 | K>N | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1436316738 CA357950771 |
78 | L>V | No |
ClinGen gnomAD |
|
|
CA3024539 rs756328481 |
81 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1328427827 COSM1049532 CA357950733 |
83 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs111452004 CA102977884 |
96 | F>L | No |
ClinGen Ensembl |
|
|
rs1241684644 CA357950564 |
105 | G>R | No |
ClinGen gnomAD |
|
|
rs1312316615 CA357950537 |
109 | A>V | No |
ClinGen gnomAD |
|
|
CA357950532 rs1560606679 |
110 | N>S | No |
ClinGen Ensembl |
|
|
CA3024514 rs778380450 |
112 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs758954731 CA3024513 |
112 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs866354491 CA102977883 |
117 | G>E | No |
ClinGen Ensembl |
|
|
rs376535772 CA102977881 |
119 | Y>C | No |
ClinGen ESP |
|
|
rs1385558748 CA357950377 |
130 | V>L | No |
ClinGen gnomAD |
|
|
CA3024495 rs758904818 |
137 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA357950307 rs1401883929 |
140 | Y>C | No |
ClinGen gnomAD |
|
|
rs748623959 CA3024494 |
142 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA357950289 rs1477226914 |
143 | T>A | No |
ClinGen gnomAD |
|
|
rs1323355157 CA357950285 |
143 | T>I | No |
ClinGen gnomAD |
|
|
COSM1049531 CA102976791 rs1000796769 |
165 | E>K | endometrium [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA3024468 rs777415869 |
173 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024467 rs142836504 |
173 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3024469 rs777415869 |
173 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024465 rs765118167 |
174 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs759365658 CA3024464 |
176 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024463 rs138929276 |
179 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1349802521 CA357950004 |
181 | A>G | No |
ClinGen gnomAD |
|
|
CA357949989 rs1441171271 |
183 | D>E | No |
ClinGen gnomAD |
|
|
rs1391497881 CA357949958 |
188 | A>G | No |
ClinGen gnomAD |
|
|
CA357949960 rs1330417912 |
188 | A>S | No |
ClinGen gnomAD |
|
|
CA357949952 rs1172335185 |
189 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1483715092 CA357949943 |
191 | M>L | No |
ClinGen TOPMed |
|
|
rs1182750511 CA357949859 |
202 | L>F | No |
ClinGen gnomAD |
|
|
CA3024456 rs769123247 |
206 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749789772 CA3024455 |
208 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1409640610 CA357949801 |
210 | D>Y | No |
ClinGen TOPMed |
|
|
rs770369705 CA3024453 |
211 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA3024430 rs778643523 |
215 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1475577978 CA357949391 |
216 | D>N | No |
ClinGen gnomAD |
|
|
rs1405078762 CA357949371 |
218 | F>L | No |
ClinGen TOPMed |
|
|
CA3024429 rs754775984 |
219 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA357949354 rs1184839287 |
221 | P>S | No |
ClinGen gnomAD |
|
|
rs1243748825 CA357949316 |
227 | M>V | No |
ClinGen gnomAD |
|
|
rs756099128 CA3024426 |
236 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357949213 rs1467830676 |
241 | N>S | No |
ClinGen gnomAD |
|
|
rs749733697 CA102976202 |
246 | E>Q | No |
ClinGen Ensembl |
|
|
rs1033778784 CA102976201 |
249 | T>S | No |
ClinGen TOPMed |
|
|
rs1270922247 CA357949150 |
250 | H>D | No |
ClinGen gnomAD |
|
|
rs1450390683 CA357949129 |
253 | V>I | No |
ClinGen TOPMed |
|
|
rs867979520 CA102976200 |
261 | S>N | No |
ClinGen gnomAD |
|
|
CA3024410 rs748997466 |
263 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1269263876 CA357948485 |
271 | H>D | No |
ClinGen TOPMed |
|
|
rs1444501462 CA357948480 |
271 | H>Q | No |
ClinGen gnomAD |
|
|
rs1355122946 CA357948474 |
272 | N>S | No |
ClinGen gnomAD |
|
|
rs1434092585 CA357948443 |
276 | S>C | No |
ClinGen gnomAD |
|
|
rs540250871 CA102975023 |
277 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024405 rs540250871 |
277 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs185115989 CA3024401 |
286 | A>G | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs542649732 CA3024402 |
286 | A>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs185115989 CA102975022 |
286 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1485755431 CA357949784 |
289 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1259403610 CA553702566 |
291 | Y>* | No |
ClinGen gnomAD |
|
|
CA357949701 rs1313190046 |
295 | Q>E | No |
ClinGen TOPMed |
|
|
rs1427394223 CA357949689 |
296 | T>S | No |
ClinGen gnomAD |
|
|
CA3024384 rs752862238 |
305 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1578427507 CA357949548 |
310 | N>D | No |
ClinGen Ensembl |
|
|
rs959612309 CA102974762 |
312 | L>I | No |
ClinGen TOPMed |
|
|
rs4146577 CA102974761 |
318 | K>Q | No |
ClinGen Ensembl |
|
|
CA357949048 rs1163377552 |
320 | A>T | No |
ClinGen gnomAD |
|
|
CA3024360 rs768159553 |
321 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA357949041 rs768159553 |
321 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1223403393 CA357948976 |
329 | M>I | No |
ClinGen TOPMed |
|
|
CA357948980 rs1450495357 |
329 | M>T | No |
ClinGen gnomAD |
|
|
rs752223096 CA3024358 |
335 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357948847 rs1209744818 |
347 | M>T | No |
ClinGen gnomAD |
|
|
CA3024355 rs776210956 |
348 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357948838 rs1337481113 |
348 | D>V | No |
ClinGen gnomAD |
|
|
CA3024353 rs760412015 |
351 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA357948765 rs1578413360 |
359 | E>G | No |
ClinGen Ensembl |
|
|
rs1409876017 CA357948678 |
370 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
CA357948668 rs1375394757 |
371 | I>S | No |
ClinGen gnomAD |
|
|
rs1403289502 CA357948617 |
379 | S>A | No |
ClinGen gnomAD |
|
|
CA3024332 rs774161035 |
383 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3024331 rs768724533 |
384 | F>C | No |
ClinGen ExAC gnomAD |
|
|
CA3024307 rs536640678 |
387 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs569765595 CA3024306 |
388 | T>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA3024305 rs776993681 |
393 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA357948307 rs1270077332 |
395 | V>G | No |
ClinGen gnomAD |
|
|
rs1201745966 CA357948297 |
397 | R>G | No |
ClinGen gnomAD |
|
|
rs1429103068 CA357948290 |
398 | N>H | No |
ClinGen gnomAD |
|
|
rs1727467178 RCV001216094 |
406 | M>I | No |
ClinVar dbSNP |
|
|
rs1436739036 CA357948231 |
406 | M>K | No |
ClinGen gnomAD |
|
|
rs769247405 CA3024273 |
419 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA357948125 rs1303693414 |
419 | S>N | No |
ClinGen gnomAD |
|
|
CA3024272 COSM3824844 rs745416220 |
422 | T>M | breast [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1372357775 CA357948093 |
425 | G>S | No |
ClinGen gnomAD |
|
|
rs1560570546 RCV000762100 |
426 | L>missing | No |
ClinVar dbSNP |
|
|
CA357948059 rs1308518859 |
430 | G>D | No |
ClinGen gnomAD |
|
|
rs1175529557 CA357948055 |
431 | M>V | No |
ClinGen TOPMed |
|
|
CA357948028 rs369318194 |
434 | S>R | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs746729174 CA3024269 |
441 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1179419824 CA357947979 |
442 | Q>L | No |
ClinGen gnomAD |
|
|
CA357947954 rs1473264145 |
446 | S>N | No |
ClinGen gnomAD |
|
|
CA357947906 rs1330073156 |
451 | A>V | No |
ClinGen gnomAD |
|
|
rs749840124 CA3024251 |
452 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763442265 CA3024233 |
464 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
RCV001008246 rs1578382902 |
465 | H>missing | No |
ClinVar dbSNP |
|
|
CA357947798 rs1374390478 |
465 | H>R | No |
ClinGen gnomAD |
|
|
CA357947770 rs1160055643 |
469 | S>T | No |
ClinGen TOPMed |
|
|
CA3024232 rs775976578 |
472 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs368195162 CA102969036 |
478 | R>Q | No |
ClinGen ESP TOPMed |
|
|
rs1360040556 CA357947673 |
483 | M>V | No |
ClinGen TOPMed |
|
|
rs867473896 CA102969035 |
484 | P>S | No |
ClinGen Ensembl |
|
|
CA357947660 rs1394307680 |
485 | P>T | No |
ClinGen gnomAD |
|
|
CA357947651 rs1296940907 |
486 | R>H | No |
ClinGen TOPMed |
|
|
CA102969033 rs878969404 |
490 | M>I | No |
ClinGen Ensembl |
|
|
rs1373331738 CA357947615 |
491 | P>R | No |
ClinGen TOPMed |
|
|
rs771722172 CA3024228 |
492 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA102969032 rs1003181111 |
494 | A>D | No |
ClinGen TOPMed gnomAD |
|
|
CA357947599 rs778848431 |
494 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM166947 CA3024226 rs778848431 |
494 | A>T | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs768576482 CA3024225 |
495 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs867409069 CA102969031 |
496 | L>F | No |
ClinGen gnomAD |
|
|
CA357947589 rs867409069 |
496 | L>I | No |
ClinGen gnomAD |
|
|
rs1230679427 CA357947569 |
499 | I>V | No |
ClinGen gnomAD |
|
|
CA357947556 rs1332460277 |
500 | N>K | No |
ClinGen gnomAD |
|
|
rs1578382769 CA357947559 |
500 | N>S | No |
ClinGen Ensembl |
|
|
CA357947532 rs1390292432 |
504 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
CA357947531 rs1390292432 |
504 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
CA357947524 rs1369276872 |
505 | S>L | No |
ClinGen gnomAD |
|
|
rs1343836557 CA357947519 |
506 | E>G | No |
ClinGen TOPMed gnomAD |
|
|
rs374275483 CA3024223 |
507 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs750488827 CA3024221 |
509 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs781181414 CA3024220 |
510 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781181414 CA357947494 |
510 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1171177832 CA357947495 |
510 | T>S | No |
ClinGen gnomAD |
|
|
rs751843872 CA3024218 |
512 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA3024217 rs199874915 |
513 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA357947467 rs1184186677 |
514 | G>A | No |
ClinGen gnomAD |
|
|
CA357947437 rs1277738384 |
518 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
CA3024215 rs753154536 |
520 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1578382624 CA357947414 |
521 | Q>R | No |
ClinGen Ensembl |
No associated diseases with Q08209
15 regional properties for Q08209
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | CRIB domain | 1585 - 1620 | IPR000095 |
| domain | Protein kinase domain | 76 - 342 | IPR000719 |
| domain | AGC-kinase, C-terminal | 343 - 413 | IPR000961 |
| domain | Citron homology (CNH) domain | 1240 - 1521 | IPR001180 |
| domain | Pleckstrin homology domain | 1096 - 1217 | IPR001849 |
| domain | Protein kinase C-like, phorbol ester/diacylglycerol-binding domain | 1026 - 1077 | IPR002219 |
| active_site | Serine/threonine-protein kinase, active site | 196 - 208 | IPR008271 |
| domain | Myotonic dystrophy protein kinase, coiled coil | 878 - 939 | IPR014930 |
| binding_site | Protein kinase, ATP binding site | 82 - 105 | IPR017441 |
| domain | Diacylglycerol/phorbol-ester binding | 1024 - 1038 | IPR020454-1 |
| domain | Diacylglycerol/phorbol-ester binding | 1040 - 1049 | IPR020454-2 |
| domain | Diacylglycerol/phorbol-ester binding | 1053 - 1064 | IPR020454-3 |
| domain | Diacylglycerol/phorbol-ester binding | 1065 - 1077 | IPR020454-4 |
| domain | KELK-motif containing domain | 528 - 606 | IPR031597 |
| domain | Serine/threonine-protein kinase MRCK beta, catalytic domain | 3 - 411 | IPR042718 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.3.16 | Phosphoric monoester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR45673 | SERINE/THREONINE-PROTEIN PHOSPHATASE 2B CATALYTIC SUBUNIT 1-RELATED |
| PANTHER Subfamily | PTHR45673:SF5 | PROTEIN PHOSPHATASE 3 CATALYTIC SUBUNIT ALPHA |
| PANTHER Protein Class |
protein phosphatase
protein modifying enzyme |
|
| PANTHER Pathway Category |
CCKR signaling map CaN T cell activation Calcineurin Wnt signaling pathway Calcineurin B cell activation Calcineurin Gonadotropin-releasing hormone receptor pathway Caln |
|
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| calcineurin complex | A heterodimeric calcium ion and calmodulin dependent protein phosphatase composed of catalytic and regulatory subunits; the regulatory subunit is very similar in sequence to calmodulin. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| extrinsic component of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to one of its surfaces, but not integrated into the hydrophobic region. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| sarcolemma | The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers. |
| Schaffer collateral - CA1 synapse | A synapse between the Schaffer collateral axon of a CA3 pyramidal cell and a CA1 pyramidal cell. |
| slit diaphragm | A specialized cell-cell junction found between the interdigitating foot processes of the glomerular epithelium (the podocytes) in the vertebrate kidney, which is adapted for facilitating glomerular filtration. |
| Z disc | Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATPase binding | Binding to an ATPase, any enzyme that catalyzes the hydrolysis of ATP. |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| calmodulin-dependent protein phosphatase activity | Catalysis of the reaction: protein serine/threonine phosphate + H2O = protein serine/threonine + phosphate, dependent on the presence of calcium-bound calmodulin. |
| cyclosporin A binding | Binding to cyclosporin A, a cyclic undecapeptide that contains several N-methylated and unusual amino acids. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| protein dimerization activity | The formation of a protein dimer, a macromolecular structure consists of two noncovalently associated identical or nonidentical subunits. |
| protein serine/threonine phosphatase activity | Catalysis of the reaction: protein serine phosphate + H2O = protein serine + phosphate, and protein threonine phosphate + H2O = protein threonine + phosphate. |
| protein-containing complex binding | Binding to a macromolecular complex. |
33 GO annotations of biological process
| Name | Definition |
|---|---|
| aging | A developmental process that is a deterioration and loss of function over time. Aging includes loss of functions such as resistance to disease, homeostasis, and fertility, as well as wear and tear. Aging includes cellular senescence, but is more inclusive. May precede death and may succeed developmental maturation (GO:0021700). |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| calcineurin-mediated signaling | Any intracellular signal transduction in which the signal is passed on within the cell by activation of a transcription factor as a consequence of dephosphorylation by Ca(2+)-activated calcineurin. The process begins with calcium-dependent activation of the phosphatase calcineurin. Calcineurin is a calcium- and calmodulin-dependent serine/threonine protein phosphatase with a conserved function in eukaryotic species from yeast to humans. In yeast and fungi, calcineurin regulates stress signaling and cell cycle, and sporulation and virulence in pathogenic fungi. In metazoans, calcineurin is involved in cell commitment, organogenesis and organ development and immune function of T-lymphocytes. By a conserved mechanism, calcineurin phosphatase activates fungal Crz1 and mammalian NFATc by dephosphorylation and translocation of these transcription factors to the nucleus to regulate gene expression. |
| calcineurin-NFAT signaling cascade | Any intracellular signal transduction in which the signal is passed on within the cell by activation of a member of the NFAT protein family as a consequence of NFAT dephosphorylation by Ca(2+)-activated calcineurin. The cascade begins with calcium-dependent activation of the phosphatase calcineurin. Calcineurin dephosphorylates multiple phosphoserine residues on NFAT, resulting in the translocation of NFAT to the nucleus. The cascade ends with regulation of transcription by NFAT. The calcineurin-NFAT cascade lies downstream of many cell surface receptors, including G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs) that signal to mobilize calcium ions (Ca2+). |
| calcium ion transport | The directed movement of calcium (Ca) ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| cardiac muscle hypertrophy in response to stress | The physiological enlargement or overgrowth of all or part of the heart muscle due to an increase in size (not length) of individual cardiac muscle fibers, without cell division, as a result of a disturbance in organismal or cellular homeostasis. |
| cellular response to glucose stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a glucose stimulus. |
| dephosphorylation | The process of removing one or more phosphoric (ester or anhydride) residues from a molecule. |
| excitatory postsynaptic potential | A process that leads to a temporary increase in postsynaptic potential due to the flow of positively charged ions into the postsynaptic cell. The flow of ions that causes an EPSP is an excitatory postsynaptic current (EPSC) and makes it easier for the neuron to fire an action potential. |
| G1/S transition of mitotic cell cycle | The mitotic cell cycle transition by which a cell in G1 commits to S phase. The process begins with the build up of G1 cyclin-dependent kinase (G1 CDK), resulting in the activation of transcription of G1 cyclins. The process ends with the positive feedback of the G1 cyclins on the G1 CDK which commits the cell to S phase, in which DNA replication is initiated. |
| multicellular organismal response to stress | Any process that results in a change in state or activity of a multicellular organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation). |
| negative regulation of chromatin binding | Any process that stops or reduces the frequency, rate or extent of chromatin binding. Chromatin binding is the selective interaction with chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| negative regulation of dendrite morphogenesis | Any process that stops, prevents, or reduces the frequency, rate or extent of dendrite morphogenesis. |
| negative regulation of insulin secretion | Any process that stops, prevents, or reduces the frequency, rate or extent of the regulated release of insulin. |
| negative regulation of production of miRNAs involved in gene silencing by miRNA | Any process that stops, prevents or reduces the frequency, rate or extent of maturation of miRNAs. |
| peptidyl-serine dephosphorylation | The removal of phosphoric residues from peptidyl-O-phospho-L-serine to form peptidyl-serine. |
| positive regulation of cardiac muscle hypertrophy in response to stress | Any process that activates or increases the frequency, rate or extent of cardiac muscle hypertrophy in response to stress. |
| positive regulation of cell adhesion | Any process that activates or increases the frequency, rate or extent of cell adhesion. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of connective tissue replacement | Any process that activates or increases the frequency, rate or extent of connective tissue replacement. |
| positive regulation of DNA-binding transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of a transcription factor, any factor involved in the initiation or regulation of transcription. |
| positive regulation of endocytosis | Any process that activates or increases the frequency, rate or extent of endocytosis. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| postsynaptic modulation of chemical synaptic transmission | Any process, acting in the postsynapse that results in modulation of chemical synaptic transmission. |
| protein dephosphorylation | The process of removing one or more phosphoric residues from a protein. |
| protein import into nucleus | The directed movement of a protein from the cytoplasm to the nucleus. |
| response to amphetamine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an amphetamine stimulus. Amphetamines consist of a group of compounds related to alpha-methylphenethylamine. |
| response to calcium ion | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a calcium ion stimulus. |
| skeletal muscle fiber development | The process whose specific outcome is the progression of the skeletal muscle fiber over time, from its formation to the mature structure. Muscle fibers are formed by the maturation of myotubes. They can be classed as slow, intermediate/fast or fast. |
| skeletal muscle tissue regeneration | The regrowth of skeletal muscle tissue to repair injured or damaged muscle fibers in the postnatal stage. |
| T cell activation | The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific. |
| transition between fast and slow fiber | The process of conversion of fast-contracting muscle fibers to a slower character. This may involve slowing of contractile rate, slow myosin gene induction, increase in oxidative metabolic properties, altered electrophysiology and altered innervation. This process also regulates skeletal muscle adapatation. |
| wound healing | The series of events that restore integrity to a damaged tissue, following an injury. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P23287 | CNA1 | Serine/threonine-protein phosphatase 2B catalytic subunit A1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P48452 | PPP3CA | Protein phosphatase 3 catalytic subunit alpha | Bos taurus (Bovine) | SS |
| Q27889 | Pp2B-14D | Serine/threonine-protein phosphatase 2B catalytic subunit 2 | Drosophila melanogaster (Fruit fly) | SS |
| P48456 | CanA1 | Serine/threonine-protein phosphatase 2B catalytic subunit 1 | Drosophila melanogaster (Fruit fly) | SS |
| Q9VXF1 | CanA-14F | Serine/threonine-protein phosphatase 2B catalytic subunit 3 | Drosophila melanogaster (Fruit fly) | SS |
| P48454 | PPP3CC | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform | Homo sapiens (Human) | SS |
| P16298 | PPP3CB | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Homo sapiens (Human) | EV |
| P48453 | Ppp3cb | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Mus musculus (Mouse) | SS |
| P48455 | Ppp3cc | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform | Mus musculus (Mouse) | PR |
| P63328 | Ppp3ca | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Mus musculus (Mouse) | EV |
| P20651 | Ppp3cb | Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform | Rattus norvegicus (Rat) | SS |
| P63329 | Ppp3ca | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | Rattus norvegicus (Rat) | SS |
| Q0G819 | tax-6 | Serine/threonine-protein phosphatase 2B catalytic subunit | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSEPKAIDPK | LSTTDRVVKA | VPFPPSHRLT | AKEVFDNDGK | PRVDILKAHL | MKEGRLEESV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ALRIITEGAS | ILRQEKNLLD | IDAPVTVCGD | IHGQFFDLMK | LFEVGGSPAN | TRYLFLGDYV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DRGYFSIECV | LYLWALKILY | PKTLFLLRGN | HECRHLTEYF | TFKQECKIKY | SERVYDACMD |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AFDCLPLAAL | MNQQFLCVHG | GLSPEINTLD | DIRKLDRFKE | PPAYGPMCDI | LWSDPLEDFG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NEKTQEHFTH | NTVRGCSYFY | SYPAVCEFLQ | HNNLLSILRA | HEAQDAGYRM | YRKSQTTGFP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SLITIFSAPN | YLDVYNNKAA | VLKYENNVMN | IRQFNCSPHP | YWLPNFMDVF | TWSLPFVGEK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VTEMLVNVLN | ICSDDELGSE | EDGFDGATAA | ARKEVIRNKI | RAIGKMARVF | SVLREESESV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| LTLKGLTPTG | MLPSGVLSGG | KQTLQSATVE | AIEADEAIKG | FSPQHKITSF | EEAKGLDRIN |
| 490 | 500 | 510 | 520 | ||
| ERMPPRRDAM | PSDANLNSIN | KALTSETNGT | DSNGSNSSNI | Q |