Q06889
Gene name |
EGR3 (PILOT) |
Protein name |
Early growth response protein 3 |
Names |
EGR-3, Zinc finger protein pilot |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1960 |
EC number |
|
Protein Class |
|
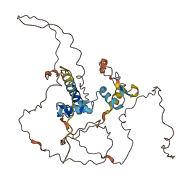
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q06889
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q06889-F1 | Predicted | AlphaFoldDB |
219 variants for Q06889
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA173884237 rs866855581 |
3 | G>D | No |
ClinGen Ensembl |
|
|
rs779115171 CA4671963 |
3 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs771517550 CA4671962 |
4 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs777860579 CA4671960 |
7 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA370566561 rs1460963748 |
8 | K>E | No |
ClinGen TOPMed |
|
|
rs756117022 CA4671959 |
8 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA173884202 rs910555755 |
10 | P>L | No |
ClinGen TOPMed |
|
|
rs748380866 CA4671958 |
11 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs1194698179 CA370566454 |
16 | L>V | No |
ClinGen gnomAD |
|
|
rs948819561 CA370566433 |
17 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA4671957 rs781530653 |
18 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs751423090 CA4671955 |
22 | D>G | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA370566199 rs1181284591 |
28 | E>D | No |
ClinGen TOPMed |
|
|
rs528695544 CA4671954 |
29 | I>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4671952 rs370975109 |
31 | S>R | No |
ClinGen ESP ExAC gnomAD |
|
|
CA370566132 rs1293727661 |
32 | A>G | No |
ClinGen TOPMed gnomAD |
|
|
CA4671951 rs764855016 |
35 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1295872629 CA370566049 |
36 | F>L | No |
ClinGen gnomAD |
|
|
rs377538095 CA370566018 |
38 | G>C | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA173884134 rs377538095 |
38 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP NCI-TCGA TOPMed gnomAD |
| TCGA novel | 40 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs763848972 CA4671948 |
41 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 41 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370565957 rs1473386867 |
41 | D>H | No |
ClinGen gnomAD |
|
| TCGA novel | 41 | D>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs760221687 CA4671947 |
45 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1451732054 CA370565839 |
47 | N>S | No |
ClinGen gnomAD |
|
|
CA4671946 rs774799132 COSM3765398 |
48 | Q>H | central_nervous_system [Cosmic] | No |
ClinGen cosmic curated ExAC |
|
rs1268832302 CA370565817 |
48 | Q>L | No |
ClinGen gnomAD |
|
|
CA4671945 rs771244734 |
51 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1206662822 CA370565006 |
53 | N>S | No |
ClinGen gnomAD |
|
|
rs1305820164 CA370564944 |
55 | M>I | No |
ClinGen gnomAD |
|
|
CA370564904 rs1272997614 |
57 | I>F | No |
ClinGen gnomAD |
|
|
rs200581271 CA173882707 |
58 | G>S | No |
ClinGen TOPMed |
|
|
rs1229393798 CA370564846 |
60 | T>A | No |
ClinGen gnomAD |
|
|
rs376367922 CA173882700 |
60 | T>N | No |
ClinGen ESP gnomAD |
|
|
rs1437360805 CA370564808 |
61 | N>T | No |
ClinGen Ensembl |
|
|
CA370564703 rs1224710321 |
65 | N>S | No |
ClinGen TOPMed |
|
|
rs777341372 COSM1456126 CA4671912 |
66 | P>Q | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1085307474 CA370564637 RCV000489532 |
68 | L>V | No |
ClinGen ClinVar dbSNP gnomAD |
|
|
rs575465744 CA4671910 |
70 | Y>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4671909 rs767220380 |
71 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA370564505 rs563408494 |
72 | G>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4671908 rs563408494 |
72 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1475019969 CA370564415 |
74 | F>C | No |
ClinGen gnomAD |
|
|
rs1413022468 CA370564366 |
76 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1430536299 CA370564337 |
77 | A>D | No |
ClinGen TOPMed |
|
|
rs371366100 CA4671905 |
78 | P>L | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1232312342 CA370564308 |
79 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs769194373 CA4671903 |
79 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1563183599 CA370564274 |
80 | N>K | No |
ClinGen Ensembl |
|
|
CA4671902 rs760733870 |
82 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA370564165 rs1364273354 |
85 | Y>C | No |
ClinGen TOPMed |
|
|
CA370564098 rs1330216076 |
88 | K>R | No |
ClinGen gnomAD |
|
|
CA4671898 rs778880883 |
92 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA370563833 rs1212497975 |
100 | D>E | No |
ClinGen TOPMed |
|
|
CA370563798 rs1371985628 |
102 | I>F | No |
ClinGen gnomAD |
|
|
CA370563788 rs1403507620 |
102 | I>S | No |
ClinGen Ensembl |
|
|
CA370563781 rs1308285447 |
103 | I>V | No |
ClinGen gnomAD |
|
|
rs1294398691 CA370563740 |
106 | M>V | No |
ClinGen TOPMed |
|
| TCGA novel | 112 | G>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs756070397 CA4671894 |
114 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA370563628 rs1325099861 |
114 | P>L | No |
ClinGen gnomAD |
|
|
rs756070397 CA4671895 |
114 | P>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 115 | P>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4671893 rs752241262 |
120 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA370563504 rs1563183498 |
122 | T>A | No |
ClinGen Ensembl |
|
|
CA370563436 rs1563183488 COSM1205064 |
124 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated Ensembl NCI-TCGA |
|
rs1242834683 CA370563387 |
126 | T>A | No |
ClinGen TOPMed |
|
|
rs1304332386 CA370563379 |
126 | T>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1242834683 CA370563392 |
126 | T>P | No |
ClinGen TOPMed |
|
| TCGA novel | 127 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370562169 rs1388998911 |
128 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
rs761224501 CA4671885 |
129 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA4671886 rs764549547 |
129 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs375652664 CA4671883 |
132 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4671882 rs759841065 |
133 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1165897488 CA370562140 |
133 | P>S | No |
ClinGen TOPMed |
|
|
CA370562116 rs1405301124 |
136 | D>E | No |
ClinGen TOPMed |
|
|
rs774657186 CA4671881 |
137 | V>M | No |
ClinGen ExAC gnomAD |
|
|
rs774703272 CA173882363 |
138 | E>K | No |
ClinGen Ensembl |
|
|
CA370562091 rs1160447609 |
140 | M>I | No |
ClinGen TOPMed |
|
|
rs998071058 CA173882357 |
142 | P>S | No |
ClinGen Ensembl |
|
|
CA370562075 rs1585222122 |
143 | A>T | No |
ClinGen Ensembl |
|
|
CA173882355 rs750955614 |
145 | P>T | No |
ClinGen gnomAD |
|
|
CA173882354 rs966229526 |
146 | P>A | No |
ClinGen TOPMed |
|
|
rs1368120293 CA370562030 |
150 | C>S | No |
ClinGen gnomAD |
|
|
rs1302668678 CA370562022 |
151 | G>C | No |
ClinGen gnomAD |
|
|
rs1363836339 CA370562016 |
152 | D>G | No |
ClinGen gnomAD |
|
|
CA173882352 rs902024202 |
152 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
rs1286855634 CA370562009 |
153 | L>F | No |
ClinGen gnomAD |
|
|
CA370562006 rs1427998996 |
153 | L>R | No |
ClinGen TOPMed |
|
| TCGA novel | 153 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1451937670 CA370561992 |
155 | S>L | No |
ClinGen gnomAD |
|
|
rs771170208 CA4671880 |
157 | P>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 158 | V>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370561971 rs1436728463 |
159 | S>P | No |
ClinGen gnomAD |
|
| TCGA novel | 160 | F>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1585222063 CA370561947 |
162 | D>A | No |
ClinGen Ensembl |
|
|
rs1287734413 CA370561943 |
162 | D>E | No |
ClinGen TOPMed |
|
| TCGA novel | 162 | D>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370561938 rs1563183321 |
163 | P>R | No |
ClinGen Ensembl |
|
|
rs369402604 CA173882317 |
164 | Q>H | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs773024571 CA4671878 |
165 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA4671877 rs769665881 |
168 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA370561905 rs1563183304 |
169 | L>V | No |
ClinGen Ensembl |
|
|
rs1585222018 CA370561898 |
170 | A>P | No |
ClinGen Ensembl |
|
|
CA4671876 rs748027200 |
171 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA370561884 rs1485726979 |
172 | S>F | No |
ClinGen gnomAD |
|
|
CA370561886 rs1015558176 |
172 | S>P | No |
ClinGen Ensembl |
|
|
rs1015558176 CA173882285 |
172 | S>T | No |
ClinGen Ensembl |
|
|
rs1485726979 CA370561883 |
172 | S>Y | No |
ClinGen gnomAD |
|
|
CA370561880 rs1237741113 |
173 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1212576969 CA370561877 |
173 | P>R | No |
ClinGen gnomAD |
|
|
CA370561879 rs1237741113 |
173 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA4671874 rs780687434 |
175 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1585221949 CA370561844 |
178 | S>P | No |
ClinGen Ensembl |
|
|
rs754431253 CA4671873 |
181 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1585221930 CA370561777 |
183 | L>S | No |
ClinGen Ensembl |
|
|
CA173882235 rs779641186 |
185 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4671871 rs779641186 |
185 | S>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201203457 CA173882210 |
189 | P>A | No |
ClinGen 1000Genomes |
|
|
rs754205481 CA4671869 |
190 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA173882204 rs571918724 |
191 | I>V | No |
ClinGen gnomAD |
|
| TCGA novel | 194 | Y>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4671867 rs376536749 |
195 | N>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4671868 rs376536749 |
195 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 196 | L>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1247449314 CA370561474 |
196 | L>V | No |
ClinGen TOPMed |
|
|
RCV001253432 rs1803935143 |
197 | Y>* | No |
ClinVar dbSNP |
|
|
rs1461943722 CA370561434 |
197 | Y>C | No |
ClinGen TOPMed |
|
|
rs1563183194 CA370561449 |
197 | Y>N | No |
ClinGen Ensembl |
|
|
CA370561377 rs1585221878 |
199 | H>P | No |
ClinGen Ensembl |
|
|
CA4671864 rs573987115 |
200 | P>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1257225292 CA370561323 CA370561317 |
201 | N>K | No |
ClinGen gnomAD |
|
|
rs1428349287 CA370561329 |
201 | N>S | No |
ClinGen gnomAD |
|
|
CA370561294 rs1189646684 |
202 | D>E | No |
ClinGen gnomAD |
|
| TCGA novel | 202 | D>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4671863 rs139824831 |
203 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs766546348 CA4671862 |
205 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs1241742304 CA370561249 |
205 | S>T | No |
ClinGen TOPMed |
|
|
rs763210546 CA4671861 |
207 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs1249606448 CA370561207 |
208 | E>* | No |
ClinGen gnomAD |
|
|
CA4671860 rs199865032 |
208 | E>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4671859 rs748093043 |
211 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA4671858 rs748093043 |
211 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA370561132 rs1172327190 |
212 | F>S | No |
ClinGen gnomAD |
|
|
CA370561053 rs1294835197 |
215 | M>I | No |
ClinGen gnomAD |
|
|
rs1585221818 CA370561033 |
216 | D>A | No |
ClinGen Ensembl |
|
|
rs1353749373 CA370560974 |
218 | I>M | No |
ClinGen gnomAD |
|
|
CA370560908 rs1392082073 |
223 | P>L | No |
ClinGen gnomAD |
|
|
rs768580665 CA4671856 |
229 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA4671855 rs746563709 |
231 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA4671854 rs779594130 |
232 | K>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 234 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370560715 rs1399425938 |
239 | I>V | No |
ClinGen TOPMed |
|
|
CA370560705 rs1157783083 |
240 | H>P | No |
ClinGen TOPMed |
|
|
rs1157783083 CA370560704 |
240 | H>R | No |
ClinGen TOPMed |
|
| TCGA novel | 241 | P>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4671851 rs778002695 |
241 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA370560663 rs1420428500 |
244 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs756487574 CA4671850 |
244 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA370560594 rs1195170524 |
249 | P>L | No |
ClinGen gnomAD |
|
|
CA370560598 rs1195170524 |
249 | P>Q | No |
ClinGen gnomAD |
|
|
rs1024939631 CA173882062 |
249 | P>T | No |
ClinGen TOPMed |
|
|
CA173882058 rs201330520 |
252 | T>S | No |
ClinGen Ensembl |
|
|
CA370560498 rs1211627273 |
257 | R>Q | No |
ClinGen gnomAD |
|
|
CA370560448 rs1585221684 |
261 | Y>S | No |
ClinGen Ensembl |
|
|
CA370560239 rs1267598020 |
271 | H>L | No |
ClinGen TOPMed |
|
|
rs1391358555 CA370560194 |
273 | R>L | No |
ClinGen gnomAD |
|
|
rs879187953 CA173882050 |
275 | H>P | No |
ClinGen Ensembl |
|
|
CA370560057 rs1205883514 |
280 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs765367375 CA4671842 |
288 | R>L | No |
ClinGen ExAC gnomAD |
|
|
CA370559913 rs1470925341 |
288 | R>S | No |
ClinGen gnomAD |
|
| TCGA novel | 289 | S>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1205700972 CA370559812 |
292 | L>V | No |
ClinGen TOPMed |
|
|
CA370559748 rs1585221564 |
295 | H>P | No |
ClinGen Ensembl |
|
| TCGA novel | 297 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370559644 rs1563182887 |
302 | H>Y | No |
ClinGen Ensembl |
|
|
CA173881955 rs903632785 |
305 | F>L | No |
ClinGen TOPMed |
|
|
rs1585221503 CA370559431 |
315 | S>N | No |
ClinGen Ensembl |
|
|
rs1563182846 CA370559313 |
321 | T>I | No |
ClinGen Ensembl |
|
|
CA370559264 rs1319333786 |
324 | I>V | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 325 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370559184 rs1256897281 |
328 | T>R | No |
ClinGen Ensembl |
|
| TCGA novel | 330 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 333 | F>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 335 | C>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs921614228 CA173881901 |
335 | C>R | No |
ClinGen TOPMed |
|
| TCGA novel | 335 | C>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA370559053 rs1322967385 |
336 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 344 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs867051535 CA173881876 |
347 | E>K | No |
ClinGen Ensembl |
|
|
rs1259984282 CA370558867 |
348 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs753655773 CA4671823 |
353 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1306826996 CA370558751 |
354 | I>V | No |
ClinGen gnomAD |
|
|
CA370558698 rs1205126454 |
356 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1392362378 CA370558691 |
356 | L>R | No |
ClinGen gnomAD |
|
|
CA4671821 rs760612686 |
357 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA4671817 rs759151796 |
363 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA370558555 rs1376507578 |
364 | E>Q | No |
ClinGen TOPMed |
|
|
COSM3432273 CA370558474 rs1171159379 |
367 | G>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs770751717 CA4671814 |
368 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4671811 rs141587813 |
374 | A>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4671810 rs201598642 |
374 | A>V | Variant assessed as Somatic; 0.0001406 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA173881755 rs768743962 |
376 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA4671808 rs768743962 |
376 | P>S | No |
ClinGen ExAC gnomAD |
|
| rs749768945 | 377 | V>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778916249 CA4671805 |
380 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA370558259 rs1233560578 |
380 | A>S | No |
ClinGen gnomAD |
|
|
rs1233560578 CA370558263 |
380 | A>T | No |
ClinGen gnomAD |
|
|
CA370558228 rs1354608257 |
382 | V>A | No |
ClinGen gnomAD |
|
|
CA370558240 rs1292913772 |
382 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs757503730 CA4671803 |
383 | V>A | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 386 | C>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs760574595 CA4671801 |
387 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760574595 CA4671800 |
387 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA370558167 rs1563182557 |
387 | A>V | No |
ClinGen Ensembl |
|
| TCGA novel | 388 | A>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
No associated diseases with Q06889
4 regional properties for Q06889
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Zinc finger C2H2-type | 275 - 304 | IPR013087-1 |
| domain | Zinc finger C2H2-type | 305 - 332 | IPR013087-2 |
| domain | Zinc finger C2H2-type | 333 - 360 | IPR013087-3 |
| domain | Early growth response, N-terminal | 87 - 173 | IPR021849 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| metal ion binding | Binding to a metal ion. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
13 GO annotations of biological process
| Name | Definition |
|---|---|
| cell migration involved in sprouting angiogenesis | The orderly movement of endothelial cells into the extracellular matrix in order to form new blood vessels involved in sprouting angiogenesis. |
| cellular response to fibroblast growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an fibroblast growth factor stimulus. |
| cellular response to vascular endothelial growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vascular endothelial growth factor stimulus. |
| circadian rhythm | Any biological process in an organism that recurs with a regularity of approximately 24 hours. |
| endothelial cell chemotaxis | The directed movement of an endothelial cell guided by a specific chemical concentration gradient. Movement may be towards a higher concentration (positive chemotaxis) or towards a lower concentration (negative chemotaxis). |
| muscle organ development | The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| neuromuscular synaptic transmission | The process of synaptic transmission from a neuron to a muscle, across a synapse. |
| peripheral nervous system development | The process whose specific outcome is the progression of the peripheral nervous system over time, from its formation to the mature structure. The peripheral nervous system is one of the two major divisions of the nervous system. Nerves in the PNS connect the central nervous system (CNS) with sensory organs, other organs, muscles, blood vessels and glands. |
| positive regulation of endothelial cell proliferation | Any process that activates or increases the rate or extent of endothelial cell proliferation. |
| positive regulation of T cell differentiation in thymus | Any process that activates or increases the frequency, rate or extent of T cell differentiation in the thymus. |
| regulation of gamma-delta T cell differentiation | Any process that modulates the frequency, rate or extent of gamma-delta T cell differentiation. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
177 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q08DS3 | OSR1 | Protein odd-skipped-related 1 | Bos taurus (Bovine) | PR |
| Q2VWH6 | FEZF2 | Fez family zinc finger protein 2 | Bos taurus (Bovine) | PR |
| A6QNZ0 | ZSCAN26 | Zinc finger and SCAN domain-containing protein 26 | Bos taurus (Bovine) | PR |
| A7MBI1 | ZFP69 | Zinc finger protein 69 homolog | Bos taurus (Bovine) | PR |
| Q08705 | CTCF | Transcriptional repressor CTCF | Gallus gallus (Chicken) | PR |
| O42409 | GFI1B | Zinc finger protein Gfi-1b | Gallus gallus (Chicken) | PR |
| A2T6W2 | ZNF449 | Zinc finger protein 449 | Pan troglodytes (Chimpanzee) | PR |
| Q9U405 | grau | Transcription factor grauzone | Drosophila melanogaster (Fruit fly) | PR |
| Q7K0S9 | sug | Zinc finger protein GLIS2 homolog | Drosophila melanogaster (Fruit fly) | PR |
| P20385 | Cf2 | Chorion transcription factor Cf2 | Drosophila melanogaster (Fruit fly) | PR |
| Q86P48 | ATbp | AT-rich binding protein | Drosophila melanogaster (Fruit fly) | PR |
| P28698 | MZF1 | Myeloid zinc finger 1 | Homo sapiens (Human) | PR |
| Q9NTW7 | ZFP64 | Zinc finger protein 64 | Homo sapiens (Human) | PR |
| O14978 | ZNF263 | Zinc finger protein 263 | Homo sapiens (Human) | PR |
| O60304 | ZNF500 | Zinc finger protein 500 | Homo sapiens (Human) | PR |
| P08151 | GLI1 | Zinc finger protein GLI1 | Homo sapiens (Human) | PR |
| Q9UFB7 | ZBTB47 | Zinc finger and BTB domain-containing protein 47 | Homo sapiens (Human) | PR |
| Q9UNY5 | ZNF232 | Zinc finger protein 232 | Homo sapiens (Human) | PR |
| Q96SZ4 | ZSCAN10 | Zinc finger and SCAN domain-containing protein 10 | Homo sapiens (Human) | PR |
| P17028 | ZNF24 | Zinc finger protein 24 | Homo sapiens (Human) | PR |
| P25490 | YY1 | Transcriptional repressor protein YY1 | Homo sapiens (Human) | SS |
| O43296 | ZNF264 | Zinc finger protein 264 | Homo sapiens (Human) | PR |
| P49711 | CTCF | Transcriptional repressor CTCF | Homo sapiens (Human) | PR |
| Q9NQX1 | PRDM5 | PR domain zinc finger protein 5 | Homo sapiens (Human) | PR |
| Q9HBE1 | PATZ1 | POZ-, AT hook-, and zinc finger-containing protein 1 | Homo sapiens (Human) | PR |
| Q8TAX0 | OSR1 | Protein odd-skipped-related 1 | Homo sapiens (Human) | PR |
| Q9UL58 | ZNF215 | Zinc finger protein 215 | Homo sapiens (Human) | PR |
| Q8TBJ5 | FEZF2 | Fez family zinc finger protein 2 | Homo sapiens (Human) | PR |
| Q96SR6 | ZNF382 | Zinc finger protein 382 | Homo sapiens (Human) | PR |
| Q96IT1 | ZNF496 | Zinc finger protein 496 | Homo sapiens (Human) | PR |
| Q96N95 | ZNF396 | Zinc finger protein 396 | Homo sapiens (Human) | PR |
| Q9ULJ3 | ZBTB21 | Zinc finger and BTB domain-containing protein 21 | Homo sapiens (Human) | PR |
| Q9H9D4 | ZNF408 | Zinc finger protein 408 | Homo sapiens (Human) | PR |
| Q13127 | REST | RE1-silencing transcription factor | Homo sapiens (Human) | PR |
| Q8IZM8 | ZNF654 | Zinc finger protein 654 | Homo sapiens (Human) | PR |
| Q14526 | HIC1 | Hypermethylated in cancer 1 protein | Homo sapiens (Human) | PR |
| P17022 | ZNF18 | Zinc finger protein 18 | Homo sapiens (Human) | PR |
| Q86XF7 | ZNF575 | Zinc finger protein 575 | Homo sapiens (Human) | PR |
| Q8NAM6 | ZSCAN4 | Zinc finger and SCAN domain-containing protein 4 | Homo sapiens (Human) | PR |
| Q08ER8 | ZNF543 | Zinc finger protein 543 | Homo sapiens (Human) | PR |
| P17029 | ZKSCAN1 | Zinc finger protein with KRAB and SCAN domains 1 | Homo sapiens (Human) | PR |
| Q8N680 | ZBTB2 | Zinc finger and BTB domain-containing protein 2 | Homo sapiens (Human) | PR |
| O95625 | ZBTB11 | Zinc finger and BTB domain-containing protein 11 | Homo sapiens (Human) | PR |
| Q9NPC7 | MYNN | Myoneurin | Homo sapiens (Human) | PR |
| Q96BV0 | ZNF775 | Zinc finger protein 775 | Homo sapiens (Human) | PR |
| Q8NF99 | ZNF397 | Zinc finger protein 397 | Homo sapiens (Human) | PR |
| Q63HK3 | ZKSCAN2 | Zinc finger protein with KRAB and SCAN domains 2 | Homo sapiens (Human) | PR |
| Q5FWF6 | ZNF789 | Zinc finger protein 789 | Homo sapiens (Human) | PR |
| Q15776 | ZKSCAN8 | Zinc finger protein with KRAB and SCAN domains 8 | Homo sapiens (Human) | PR |
| Q53GI3 | ZNF394 | Zinc finger protein 394 | Homo sapiens (Human) | PR |
| O95125 | ZNF202 | Zinc finger protein 202 | Homo sapiens (Human) | PR |
| Q05516 | ZBTB16 | Zinc finger and BTB domain-containing protein 16 | Homo sapiens (Human) | PR |
| Q9H116 | GZF1 | GDNF-inducible zinc finger protein 1 | Homo sapiens (Human) | PR |
| Q8N0Y2 | ZNF444 | Zinc finger protein 444 | Homo sapiens (Human) | PR |
| Q6P9G9 | ZNF449 | Zinc finger protein 449 | Homo sapiens (Human) | PR |
| Q8IW36 | ZNF695 | Zinc finger protein 695 | Homo sapiens (Human) | PR |
| Q5VTD9 | GFI1B | Zinc finger protein Gfi-1b | Homo sapiens (Human) | PR |
| Q6PG37 | ZNF790 | Zinc finger protein 790 | Homo sapiens (Human) | PR |
| Q9NQV6 | PRDM10 | PR domain zinc finger protein 10 | Homo sapiens (Human) | PR |
| Q9Y2D9 | ZNF652 | Zinc finger protein 652 | Homo sapiens (Human) | PR |
| Q5TC79 | ZBTB37 | Zinc finger and BTB domain-containing protein 37 | Homo sapiens (Human) | PR |
| Q9Y4E5 | ZNF451 | E3 SUMO-protein ligase ZNF451 | Homo sapiens (Human) | PR |
| Q8ND82 | ZNF280C | Zinc finger protein 280C | Homo sapiens (Human) | PR |
| Q49AA0 | ZFP69 | Zinc finger protein 69 homolog | Homo sapiens (Human) | PR |
| O43298 | ZBTB43 | Zinc finger and BTB domain-containing protein 43 | Homo sapiens (Human) | PR |
| Q9Y330 | ZBTB12 | Zinc finger and BTB domain-containing protein 12 | Homo sapiens (Human) | PR |
| Q13105 | ZBTB17 | Zinc finger and BTB domain-containing protein 17 | Homo sapiens (Human) | PR |
| P51508 | ZNF81 | Zinc finger protein 81 | Homo sapiens (Human) | PR |
| Q5JNZ3 | ZNF311 | Zinc finger protein 311 | Homo sapiens (Human) | PR |
| Q9BRR0 | ZKSCAN3 | Zinc finger protein with KRAB and SCAN domains 3 | Homo sapiens (Human) | PR |
| Q969J2 | ZKSCAN4 | Zinc finger protein with KRAB and SCAN domains 4 | Homo sapiens (Human) | PR |
| P49910 | ZNF165 | Zinc finger protein 165 | Homo sapiens (Human) | PR |
| P10074 | ZBTB48 | Telomere zinc finger-associated protein | Homo sapiens (Human) | PR |
| P17010 | ZFX | Zinc finger X-chromosomal protein | Homo sapiens (Human) | PR |
| Q9H5H4 | ZNF768 | Zinc finger protein 768 | Homo sapiens (Human) | PR |
| Q6NSZ9 | ZSCAN25 | Zinc finger and SCAN domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q9Y2L8 | ZKSCAN5 | Zinc finger protein with KRAB and SCAN domains 5 | Homo sapiens (Human) | PR |
| Q86UZ6 | ZBTB46 | Zinc finger and BTB domain-containing protein 46 | Homo sapiens (Human) | PR |
| Q9NX65 | ZSCAN32 | Zinc finger and SCAN domain-containing protein 32 | Homo sapiens (Human) | PR |
| O14771 | ZNF213 | Zinc finger protein 213 | Homo sapiens (Human) | PR |
| Q8IWY8 | ZSCAN29 | Zinc finger and SCAN domain-containing protein 29 | Homo sapiens (Human) | PR |
| Q8NCP5 | ZBTB44 | Zinc finger and BTB domain-containing protein 44 | Homo sapiens (Human) | PR |
| P41182 | BCL6 | B-cell lymphoma 6 protein | Homo sapiens (Human) | PR |
| Q9NQX0 | PRDM6 | Putative histone-lysine N-methyltransferase PRDM6 | Homo sapiens (Human) | PR |
| Q9BU19 | ZNF692 | Zinc finger protein 692 | Homo sapiens (Human) | PR |
| Q08AG5 | ZNF844 | Zinc finger protein 844 | Homo sapiens (Human) | PR |
| Q6R2W3 | ZBED9 | SCAN domain-containing protein 3 | Homo sapiens (Human) | PR |
| P98182 | ZNF200 | Zinc finger protein 200 | Homo sapiens (Human) | PR |
| Q9UK11 | ZNF223 | Zinc finger protein 223 | Homo sapiens (Human) | PR |
| O15156 | ZBTB7B | Zinc finger and BTB domain-containing protein 7B | Homo sapiens (Human) | PR |
| Q6ZMS7 | ZNF783 | Zinc finger protein 783 | Homo sapiens (Human) | PR |
| P59923 | ZNF445 | Zinc finger protein 445 | Homo sapiens (Human) | PR |
| Q8N859 | ZNF713 | Zinc finger protein 713 | Homo sapiens (Human) | PR |
| Q8TD17 | ZNF398 | Zinc finger protein 398 | Homo sapiens (Human) | PR |
| P52739 | ZNF131 | Zinc finger protein 131 | Homo sapiens (Human) | PR |
| A6NGD5 | ZSCAN5C | Zinc finger and SCAN domain-containing protein 5C | Homo sapiens (Human) | PR |
| Q7Z398 | ZNF550 | Zinc finger protein 550 | Homo sapiens (Human) | PR |
| Q9Y2K1 | ZBTB1 | Zinc finger and BTB domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q96N20 | ZNF75A | Zinc finger protein 75A | Homo sapiens (Human) | PR |
| A6NJL1 | ZSCAN5B | Zinc finger and SCAN domain-containing protein 5B | Homo sapiens (Human) | PR |
| A1YPR0 | ZBTB7C | Zinc finger and BTB domain-containing protein 7C | Homo sapiens (Human) | PR |
| Q9NWS9 | ZNF446 | Zinc finger protein 446 | Homo sapiens (Human) | PR |
| P24278 | ZBTB25 | Zinc finger and BTB domain-containing protein 25 | Homo sapiens (Human) | PR |
| Q96N38 | ZNF714 | Zinc finger protein 714 | Homo sapiens (Human) | PR |
| Q86YH2 | ZNF280B | Zinc finger protein 280B | Homo sapiens (Human) | PR |
| Q9Y5W3 | KLF2 | Krueppel-like factor 2 | Homo sapiens (Human) | PR |
| P18146 | EGR1 | Early growth response protein 1 | Homo sapiens (Human) | PR |
| Q05215 | EGR4 | Early growth response protein 4 | Homo sapiens (Human) | PR |
| Q9Y4X4 | KLF12 | Krueppel-like factor 12 | Homo sapiens (Human) | PR |
| Q99612 | KLF6 | Krueppel-like factor 6 | Homo sapiens (Human) | PR |
| O75840 | KLF7 | Krueppel-like factor 7 | Homo sapiens (Human) | PR |
| P57682 | KLF3 | Krueppel-like factor 3 | Homo sapiens (Human) | PR |
| O08584 | Klf6 | Krueppel-like factor 6 | Mus musculus (Mouse) | PR |
| Q61164 | Ctcf | Transcriptional repressor CTCF | Mus musculus (Mouse) | PR |
| Q810A1 | Znf18 | Zinc finger protein 18 | Mus musculus (Mouse) | PR |
| Q8BGS3 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Mus musculus (Mouse) | PR |
| Q00899 | Yy1 | Transcriptional repressor protein YY1 | Mus musculus (Mouse) | PR |
| P41183 | Bcl6 | B-cell lymphoma 6 protein homolog | Mus musculus (Mouse) | PR |
| Q9DAI4 | Zbtb43 | Zinc finger and BTB domain-containing protein 43 | Mus musculus (Mouse) | PR |
| O70237 | Gfi1b | Zinc finger protein Gfi-1b | Mus musculus (Mouse) | PR |
| Q99KZ6 | Znf639 | Zinc finger protein 639 | Mus musculus (Mouse) | PR |
| Q9Z1D9 | Znf394 | Zinc finger protein 394 | Mus musculus (Mouse) | PR |
| Q9CXE0 | Prdm5 | PR domain zinc finger protein 5 | Mus musculus (Mouse) | PR |
| Q9DAU9 | Znf654 | Zinc finger protein 654 | Mus musculus (Mouse) | PR |
| Q9R1Y5 | Hic1 | Hypermethylated in cancer 1 protein | Mus musculus (Mouse) | PR |
| Q8R0T2 | Znf768 | Zinc finger protein 768 | Mus musculus (Mouse) | PR |
| Q9WVG7 | Osr1 | Protein odd-skipped-related 1 | Mus musculus (Mouse) | PR |
| Q8BI73 | Znf775 | Zinc finger protein 775 | Mus musculus (Mouse) | PR |
| Q8VCZ7 | Zbtb7c | Zinc finger and BTB domain-containing protein 7C | Mus musculus (Mouse) | PR |
| Q91VN1 | Znf24 | Zinc finger protein 24 | Mus musculus (Mouse) | PR |
| Q9DB38 | Znf580 | Zinc finger protein 580 | Mus musculus (Mouse) | PR |
| A7KBS4 | Zscan4d | Zinc finger and SCAN domain containing protein 4D | Mus musculus (Mouse) | PR |
| Q91VW9 | Zkscan3 | Zinc finger protein with KRAB and SCAN domains 3 | Mus musculus (Mouse) | PR |
| P10925 | Zfy1 | Zinc finger Y-chromosomal protein 1 | Mus musculus (Mouse) | PR |
| Q3TTC2 | Yy2 | Transcription factor YY2 | Mus musculus (Mouse) | PR |
| Q3UTQ7 | Prdm10 | PR domain zinc finger protein 10 | Mus musculus (Mouse) | PR |
| Q6P3Y5 | Znf280c | Zinc finger protein 280C | Mus musculus (Mouse) | PR |
| Q9ERU3 | Znf22 | Zinc finger protein 22 | Mus musculus (Mouse) | PR |
| Q8VIG1 | Rest | RE1-silencing transcription factor | Mus musculus (Mouse) | PR |
| Q9Z1D8 | Zkscan5 | Zinc finger protein with KRAB and SCAN domains 5 | Mus musculus (Mouse) | PR |
| Q8BID6 | Zbtb46 | Zinc finger and BTB domain-containing protein 46 | Mus musculus (Mouse) | PR |
| P17012 | Zfx | Zinc finger X-chromosomal protein | Mus musculus (Mouse) | PR |
| Q9WUK6 | Zbtb18 | Zinc finger and BTB domain-containing protein 18 | Mus musculus (Mouse) | PR |
| O35738 | Klf12 | Krueppel-like factor 12 | Mus musculus (Mouse) | PR |
| B2RXC5 | Znf382 | Zinc finger protein 382 | Mus musculus (Mouse) | PR |
| O08900 | Ikzf3 | Zinc finger protein Aiolos | Mus musculus (Mouse) | PR |
| Q5DU09 | Znf652 | Zinc finger protein 652 | Mus musculus (Mouse) | PR |
| Q5RJ54 | Zscan26 | Zinc finger and SCAN domain-containing protein 26 | Mus musculus (Mouse) | PR |
| Q8BLM0 | Klf8 | Krueppel-like factor 8 | Mus musculus (Mouse) | PR |
| Q99JB0 | Klf7 | Krueppel-like factor 7 | Mus musculus (Mouse) | PR |
| Q8R0A2 | Zbtb44 | Zinc finger and BTB domain-containing protein 44 | Mus musculus (Mouse) | PR |
| P20662 | Zfy2 | Zinc finger Y-chromosomal protein 2 | Mus musculus (Mouse) | PR |
| Q80VJ6 | Zscan4c | Zinc finger and SCAN domain containing protein 4C | Mus musculus (Mouse) | PR |
| Q3URS2 | Zscan4f | Zinc finger and SCAN domain containing protein 4F | Mus musculus (Mouse) | PR |
| Q60980 | Klf3 | Krueppel-like factor 3 | Mus musculus (Mouse) | PR |
| Q8K3J5 | Znf131 | Zinc finger protein 131 | Mus musculus (Mouse) | PR |
| P08046 | Egr1 | Early growth response protein 1 | Mus musculus (Mouse) | PR |
| P43300 | Egr3 | Early growth response protein 3 | Mus musculus (Mouse) | PR |
| Q9Z2K3 | Znf394 | Zinc finger protein 394 | Rattus norvegicus (Rat) | PR |
| Q642B9 | Znf18 | Zinc finger protein 18 | Rattus norvegicus (Rat) | PR |
| B0K011 | Osr1 | Protein odd-skipped-related 1 | Rattus norvegicus (Rat) | PR |
| D3ZUU2 | Gzf1 | GDNF-inducible zinc finger protein 1 | Rattus norvegicus (Rat) | PR |
| B1WBU4 | Zbtb8a | Zinc finger and BTB domain-containing protein 8A | Rattus norvegicus (Rat) | PR |
| Q7TNK3 | Znf24 | Zinc finger protein 24 | Rattus norvegicus (Rat) | PR |
| O35819 | Klf6 | Krueppel-like factor 6 | Rattus norvegicus (Rat) | PR |
| Q9R1D1 | Ctcf | Transcriptional repressor CTCF | Rattus norvegicus (Rat) | PR |
| P08154 | Egr1 | Early growth response protein 1 | Rattus norvegicus (Rat) | PR |
| A0JPL0 | Znf382 | Zinc finger protein 382 | Rattus norvegicus (Rat) | PR |
| Q4KLI1 | Zkscan1 | Zinc finger protein with KRAB and SCAN domains 1 | Rattus norvegicus (Rat) | PR |
| A1L1J6 | Znf652 | Zinc finger protein 652 | Rattus norvegicus (Rat) | PR |
| P43301 | Egr3 | Early growth response protein 3 | Rattus norvegicus (Rat) | PR |
| Q9SHD0 | ZAT4 | Zinc finger protein ZAT4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q0P4X6 | zbtb44 | Zinc finger and BTB domain-containing protein 44 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q6P882 | zbtb8a.2 | Zinc finger and BTB domain-containing protein 8A.2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| A4II20 | egr1 | Early growth response protein 1 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q567C6 | znf367 | Zinc finger protein 367 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| A7Y7X5 | znf711 | Zinc finger protein 711 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTGKLAEKLP | VTMSSLLNQL | PDNLYPEEIP | SALNLFSGSS | DSVVHYNQMA | TENVMDIGLT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NEKPNPELSY | SGSFQPAPGN | KTVTYLGKFA | FDSPSNWCQD | NIISLMSAGI | LGVPPASGAL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| STQTSTASMV | QPPQGDVEAM | YPALPPYSNC | GDLYSEPVSF | HDPQGNPGLA | YSPQDYQSAK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PALDSNLFPM | IPDYNLYHHP | NDMGSIPEHK | PFQGMDPIRV | NPPPITPLET | IKAFKDKQIH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PGFGSLPQPP | LTLKPIRPRK | YPNRPSKTPL | HERPHACPAE | GCDRRFSRSD | ELTRHLRIHT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GHKPFQCRIC | MRSFSRSDHL | TTHIRTHTGE | KPFACEFCGR | KFARSDERKR | HAKIHLKQKE |
| 370 | 380 | ||||
| KKAEKGGAPS | ASSAPPVSLA | PVVTTCA |