Q05609
Gene name |
CTR1 |
Protein name |
Serine/threonine-protein kinase CTR1 |
Names |
Protein CONSTITUTIVE TRIPLE RESPONSE1 |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G03730 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
693-701 (Activation loop from InterPro)
Target domain |
551-809 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
712-716 (Activation loop from InterPro)
Target domain |
551-809 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
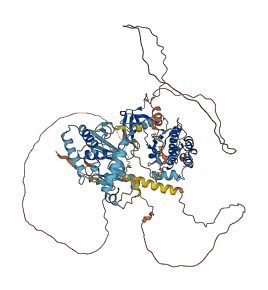
Autoinhibited structure

Activated structure

3 structures for Q05609
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3P86 | X-ray | 250 A | A/B | 540-821 | PDB |
| 3PPZ | X-ray | 299 A | A/B | 540-821 | PDB |
| AF-Q05609-F1 | Predicted | AlphaFoldDB |
30 variants for Q05609
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH03017325 | 74 | N>S | No | 1000Genomes | |
| tmp_5_979422_A_G | 80 | V>A | No | 1000Genomes | |
| tmp_5_979420_C_A | 81 | G>W | No | 1000Genomes | |
| ENSVATH03017324 | 86 | A>V | No | 1000Genomes | |
| tmp_5_979328_C_T | 111 | M>I | No | 1000Genomes | |
| tmp_5_979326_G_C | 112 | P>R | No | 1000Genomes | |
| ENSVATH06917453 | 117 | A>T | No | 1000Genomes | |
| tmp_5_979308_G_A | 118 | A>V | No | 1000Genomes | |
| ENSVATH10542190 | 127 | P>L | No | 1000Genomes | |
| ENSVATH13906810 | 144 | R>S | No | 1000Genomes | |
| ENSVATH10542189 | 150 | D>E | No | 1000Genomes | |
| tmp_5_979179_C_G | 161 | W>S | No | 1000Genomes | |
| ENSVATH13906808 | 204 | S>W | No | 1000Genomes | |
| ENSVATH00604903 | 245 | C>S | No | 1000Genomes | |
| tmp_5_978570_C_A | 306 | D>Y | No | 1000Genomes | |
| ENSVATH13906703 | 322 | M>T | No | 1000Genomes | |
| ENSVATH10542069 | 381 | K>R | No | 1000Genomes | |
| tmp_5_977737_G_A | 434 | P>S | No | 1000Genomes | |
| ENSVATH13906702 | 437 | K>E | No | 1000Genomes | |
| ENSVATH13906699 | 492 | N>Y | No | 1000Genomes | |
| ENSVATH13906698 | 510 | R>H | No | 1000Genomes | |
| tmp_5_977117_A_T | 521 | N>K | No | 1000Genomes | |
| ENSVATH06917437 | 522 | A>T | No | 1000Genomes | |
| tmp_5_977103_C_T | 526 | S>N | No | 1000Genomes | |
| tmp_5_977025_T_A | 552 | N>I | No | 1000Genomes | |
| ENSVATH06917424 | 644 | E>D | No | 1000Genomes | |
| tmp_5_975622_C_T | 699 | R>Q | No | 1000Genomes | |
| ENSVATH06917413 | 727 | P>L | No | 1000Genomes | |
| tmp_5_974978_G_C | 816 | P>A | No | 1000Genomes | |
| tmp_5_974971_C_T | 818 | R>H | No | 1000Genomes |
No associated diseases with Q05609
4 regional properties for Q05609
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 551 - 809 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 551 - 804 | IPR001245 |
| active_site | Serine/threonine-protein kinase, active site | 672 - 684 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 557 - 578 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| ethylene-activated signaling pathway | The series of molecular signals generated by the reception of ethylene (ethene, C2H4) by a receptor and ending with modulation of a cellular process, e.g. transcription. |
| gibberellin biosynthetic process | The chemical reactions and pathways resulting in the formation of gibberellin. Gibberellins are a class of highly modified terpenes that function as plant growth regulators. |
| negative regulation of ethylene-activated signaling pathway | Any process that stops or prevents ethylene (ethene) signal transduction. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| regulation of post-embryonic root development | Any process that modulates the frequency, rate or extent of post-embryonic root development. |
| regulation of stem cell division | Any process that modulates the frequency, rate or extent of stem cell division. |
| regulation of timing of transition from vegetative to reproductive phase | The process controlling the point in time during development when a vegetative meristem will change its identity to become an inflorescence or floral meristem, and/or the rate at which the change occurs. |
| response to ethylene | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ethylene (ethene) stimulus. |
| response to fructose | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a fructose stimulus. |
| response to hypoxia | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| response to sucrose | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a sucrose stimulus. |
| sugar mediated signaling pathway | The process in which a change in the level of a mono- or disaccharide such as glucose, fructose or sucrose triggers the expression of genes controlling metabolic and developmental processes. |
55 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VDU3 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Bos taurus (Bovine) | SS |
| Q4TVR5 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Bos taurus (Bovine) | PR |
| A7E3S4 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | Bos taurus (Bovine) | SS |
| Q3SZJ2 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Bos taurus (Bovine) | PR |
| Q6XUX0 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Gallus gallus (Chicken) | PR |
| P05625 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | Gallus gallus (Chicken) | PR |
| Q04982 | BRAF | Serine/threonine-protein kinase B-raf | Gallus gallus (Chicken) | SS |
| P83104 | Takl1 | Putative mitogen-activated protein kinase kinase kinase 7-like | Drosophila melanogaster (Fruit fly) | PR |
| P11346 | Raf | Raf homolog serine/threonine-protein kinase Raf | Drosophila melanogaster (Fruit fly) | PR |
| Q95UN8 | slpr | Mitogen-activated protein kinase kinase kinase | Drosophila melanogaster (Fruit fly) | EV |
| Q8NB16 | MLKL | Mixed lineage kinase domain-like protein | Homo sapiens (Human) | EV |
| P00540 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Homo sapiens (Human) | PR |
| Q5TCX8 | MAP3K21 | Mitogen-activated protein kinase kinase kinase 21 | Homo sapiens (Human) | PR |
| Q6XUX3 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Homo sapiens (Human) | PR |
| O43318 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Homo sapiens (Human) | SS |
| Q9NYL2 | MAP3K20 | Mitogen-activated protein kinase kinase kinase 20 | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| P04049 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| P10398 | ARAF | Serine/threonine-protein kinase A-Raf | Homo sapiens (Human) | PR |
| Q02779 | MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | Homo sapiens (Human) | SS |
| P15056 | BRAF | Serine/threonine-protein kinase B-raf | Homo sapiens (Human) | EV |
| Q13418 | ILK | Integrin-linked protein kinase | Homo sapiens (Human) | PR |
| Q16584 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | Homo sapiens (Human) | EV |
| O43353 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Homo sapiens (Human) | PR |
| P80192 | MAP3K9 | Mitogen-activated protein kinase kinase kinase 9 | Homo sapiens (Human) | SS |
| Q62073 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Mus musculus (Mouse) | EV |
| Q9D2Y4 | Mlkl | Mixed lineage kinase domain-like protein | Mus musculus (Mouse) | SS |
| P00536 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Mus musculus (Mouse) | PR |
| P28028 | Braf | Serine/threonine-protein kinase B-raf | Mus musculus (Mouse) | SS |
| Q99N57 | Raf1 | RAF proto-oncogene serine/threonine-protein kinase | Mus musculus (Mouse) | SS |
| P58801 | Ripk2 | Receptor-interacting serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q3U1V8 | Map3k9 | Mitogen-activated protein kinase kinase kinase 9 | Mus musculus (Mouse) | SS |
| Q66L42 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Mus musculus (Mouse) | SS |
| Q80XI6 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Mus musculus (Mouse) | PR |
| Q8VDG6 | Map3k21 | Mitogen-activated protein kinase kinase kinase 21 | Mus musculus (Mouse) | PR |
| P04627 | Araf | Serine/threonine-protein kinase A-Raf | Mus musculus (Mouse) | PR |
| O55222 | Ilk | Integrin-linked protein kinase | Mus musculus (Mouse) | PR |
| Q9ESL4 | Map3k20 | Mitogen-activated protein kinase kinase kinase 20 | Mus musculus (Mouse) | PR |
| P0C8E4 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Rattus norvegicus (Rat) | SS |
| P00539 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Rattus norvegicus (Rat) | PR |
| D3ZG83 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Rattus norvegicus (Rat) | SS |
| Q66HA1 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Rattus norvegicus (Rat) | PR |
| P11345 | Raf1 | RAF proto-oncogene serine/threonine-protein kinase | Rattus norvegicus (Rat) | SS |
| P32577 | Csk | Tyrosine-protein kinase CSK | Rattus norvegicus (Rat) | SS |
| P41243 | Matk | Megakaryocyte-associated tyrosine-protein kinase | Rattus norvegicus (Rat) | SS |
| P14056 | Araf | Serine/threonine-protein kinase A-Raf | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q07292 | lin-45 | Raf homolog serine/threonine-protein kinase | Caenorhabditis elegans | PR |
| Q9TZC4 | pat-4 | Integrin-linked protein kinase homolog pat-4 | Caenorhabditis elegans | PR |
| Q9FPR3 | EDR1 | Serine/threonine-protein kinase EDR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q67E00 | dstyk | Dual serine/threonine and tyrosine protein kinase | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEMPGRRSNY | TLLSQFSDDQ | VSVSVTGAPP | PHYDSLSSEN | RSNHNSGNTG | KAKAERGGFD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| WDPSGGGGGD | HRLNNQPNRV | GNNMYASSLG | LQRQSSGSSF | GESSLSGDYY | MPTLSAAANE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IESVGFPQDD | GFRLGFGGGG | GDLRIQMAAD | SAGGSSSGKS | WAQQTEESYQ | LQLALALRLS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SEATCADDPN | FLDPVPDESA | LRTSPSSAET | VSHRFWVNGC | LSYYDKVPDG | FYMMNGLDPY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IWTLCIDLHE | SGRIPSIESL | RAVDSGVDSS | LEAIIVDRRS | DPAFKELHNR | VHDISCSCIT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TKEVVDQLAK | LICNRMGGPV | IMGEDELVPM | WKECIDGLKE | IFKVVVPIGS | LSVGLCRHRA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LLFKVLADII | DLPCRIAKGC | KYCNRDDAAS | CLVRFGLDRE | YLVDLVGKPG | HLWEPDSLLN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GPSSISISSP | LRFPRPKPVE | PAVDFRLLAK | QYFSDSQSLN | LVFDPASDDM | GFSMFHRQYD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| NPGGENDALA | ENGGGSLPPS | ANMPPQNMMR | ASNQIEAAPM | NAPPISQPVP | NRANRELGLD |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GDDMDIPWCD | LNIKEKIGAG | SFGTVHRAEW | HGSDVAVKIL | MEQDFHAERV | NEFLREVAIM |
| 610 | 620 | 630 | 640 | 650 | 660 |
| KRLRHPNIVL | FMGAVTQPPN | LSIVTEYLSR | GSLYRLLHKS | GAREQLDERR | RLSMAYDVAK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| GMNYLHNRNP | PIVHRDLKSP | NLLVDKKYTV | KVCDFGLSRL | KASTFLSSKS | AAGTPEWMAP |
| 730 | 740 | 750 | 760 | 770 | 780 |
| EVLRDEPSNE | KSDVYSFGVI | LWELATLQQP | WGNLNPAQVV | AAVGFKCKRL | EIPRNLNPQV |
| 790 | 800 | 810 | 820 | ||
| AAIIEGCWTN | EPWKRPSFAT | IMDLLRPLIK | SAVPPPNRSD | L |