Q04206
Gene name |
RELA (NFKB3) |
Protein name |
Transcription factor p65 |
Names |
Nuclear factor NF-kappa-B p65 subunit, Nuclear factor of kappa light polypeptide gene enhancer in B-cells 3 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5970 |
EC number |
|
Protein Class |
NUCLEAR FACTOR NF-KAPPA-B PROTEIN (PTHR24169) |
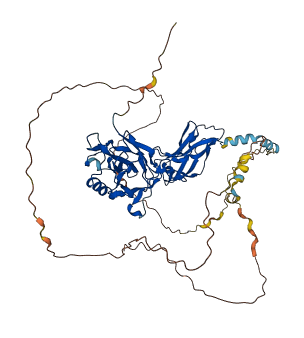
Descriptions
The inducible transcription factor NF-kB (nuclear factor kB) is responsible for regulating the expression of a wide variety of genes. The most abundant form of the protein is a heterodimer of p50 and p65 subunits, in which the p65 subunit (RELA) contains the transcriptional activation domain. Full-length RELA does not bind efficiently to DNA, but a shorter fragment lacking the C-terminal sequence binds to DNA with high affinity. Binding assay revealed that the C-terminal region interacts with the N-terminal region for the autoinhibition of N-terminal domain. The C-terminal region contains a CBP binding site and the binding of CBP to RELA enhances the transcriptional activity of RELA.
Autoinhibitory domains (AIDs)
Target domain |
1-194 (N-terminal region) |
Relief mechanism |
Partner binding, PTM |
Assay |
Deletion assay, Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

89 structures for Q04206
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1NFI | X-ray | 270 A | A/C | 20-320 | PDB |
| 2LSP | NMR | - | A | 304-316 | PDB |
| 2O61 | X-ray | 280 A | A | 20-291 | PDB |
| 3GUT | X-ray | 359 A | A/C/E/G | 20-291 | PDB |
| 3QXY | X-ray | 209 A | P/Q | 302-316 | PDB |
| 3RC0 | X-ray | 219 A | P/Q | 302-316 | PDB |
| 4KV1 | X-ray | 150 A | C/D | 308-314 | PDB |
| 4KV4 | X-ray | 200 A | B | 308-314 | PDB |
| 5U4K | NMR | - | B | 521-551 | PDB |
| 5URN | NMR | - | B | 521-551 | PDB |
| 6NV2 | X-ray | 113 A | P | 39-51 | PDB |
| 6QHL | X-ray | 120 A | P | 38-51 | PDB |
| 6QHM | X-ray | 125 A | P | 275-287 | PDB |
| 6YOW | X-ray | 123 A | P | 39-51 | PDB |
| 6YOX | X-ray | 205 A | P | 39-51 | PDB |
| 6YOY | X-ray | 180 A | P | 39-51 | PDB |
| 6YP2 | X-ray | 180 A | P | 39-51 | PDB |
| 6YP3 | X-ray | 180 A | P | 39-51 | PDB |
| 6YP8 | X-ray | 180 A | P | 39-51 | PDB |
| 6YPL | X-ray | 180 A | P | 39-51 | PDB |
| 6YPY | X-ray | 140 A | P | 39-51 | PDB |
| 6YQ2 | X-ray | 140 A | P | 39-51 | PDB |
| 7BI3 | X-ray | 120 A | P | 39-51 | PDB |
| 7BIQ | X-ray | 120 A | P | 39-51 | PDB |
| 7BIW | X-ray | 120 A | P | 39-51 | PDB |
| 7BIY | X-ray | 180 A | P | 39-51 | PDB |
| 7BJB | X-ray | 180 A | P | 39-51 | PDB |
| 7BJF | X-ray | 140 A | P | 39-51 | PDB |
| 7BJL | X-ray | 140 A | P | 39-51 | PDB |
| 7BJW | X-ray | 140 A | P | 39-51 | PDB |
| 7BKH | X-ray | 140 A | P | 39-51 | PDB |
| 7LET | X-ray | 240 A | C | 294-315 | PDB |
| 7LEU | X-ray | 282 A | B/C | 294-315 | PDB |
| 7LF4 | X-ray | 285 A | D/E | 294-315 | PDB |
| 7NJ9 | X-ray | 140 A | P | 39-51 | PDB |
| 7NJB | X-ray | 140 A | P | 39-51 | PDB |
| 7NK3 | X-ray | 140 A | P | 39-51 | PDB |
| 7NK5 | X-ray | 140 A | P | 39-51 | PDB |
| 7NLA | X-ray | 140 A | P | 39-51 | PDB |
| 7NLE | X-ray | 140 A | P | 39-51 | PDB |
| 7NM1 | X-ray | 140 A | P | 39-51 | PDB |
| 7NM3 | X-ray | 140 A | P | 39-51 | PDB |
| 7NM9 | X-ray | 170 A | P | 39-51 | PDB |
| 7NMH | X-ray | 140 A | P | 39-51 | PDB |
| 7NQP | X-ray | 124 A | P | 39-51 | PDB |
| 7NR7 | X-ray | 140 A | P | 39-51 | PDB |
| 7NSV | X-ray | 133 A | P | 39-51 | PDB |
| 7NV4 | X-ray | 120 A | P | 39-51 | PDB |
| 7NVI | X-ray | 120 A | P | 39-51 | PDB |
| 7NWS | X-ray | 120 A | P | 39-51 | PDB |
| 7NXS | X-ray | 120 A | P | 39-51 | PDB |
| 7NXT | X-ray | 120 A | P | 39-51 | PDB |
| 7NXW | X-ray | 120 A | P | 39-51 | PDB |
| 7NXY | X-ray | 120 A | P | 39-51 | PDB |
| 7NY4 | X-ray | 140 A | P | 39-51 | PDB |
| 7NYE | X-ray | 140 A | P | 39-51 | PDB |
| 7NYF | X-ray | 140 A | P | 39-51 | PDB |
| 7NYG | X-ray | 140 A | P | 39-51 | PDB |
| 7NZ6 | X-ray | 140 A | P | 39-51 | PDB |
| 7NZG | X-ray | 140 A | P | 39-51 | PDB |
| 7NZK | X-ray | 140 A | P | 39-51 | PDB |
| 7NZV | X-ray | 140 A | P | 39-51 | PDB |
| 7O34 | X-ray | 120 A | P | 39-51 | PDB |
| 7O3A | X-ray | 120 A | P | 39-51 | PDB |
| 7O3F | X-ray | 140 A | P | 39-51 | PDB |
| 7O3P | X-ray | 140 A | P | 39-51 | PDB |
| 7O3Q | X-ray | 180 A | P | 39-51 | PDB |
| 7O3R | X-ray | 180 A | P | 39-51 | PDB |
| 7O3S | X-ray | 200 A | P | 39-51 | PDB |
| 7O57 | X-ray | 140 A | P | 39-51 | PDB |
| 7O59 | X-ray | 120 A | P | 39-51 | PDB |
| 7O5A | X-ray | 180 A | P | 39-51 | PDB |
| 7O5C | X-ray | 180 A | P | 39-51 | PDB |
| 7O5D | X-ray | 180 A | P | 39-51 | PDB |
| 7O5F | X-ray | 180 A | P | 39-51 | PDB |
| 7O5G | X-ray | 180 A | P | 39-51 | PDB |
| 7O5O | X-ray | 180 A | P | 39-51 | PDB |
| 7O5P | X-ray | 180 A | P | 39-51 | PDB |
| 7O5S | X-ray | 180 A | P | 39-51 | PDB |
| 7O5U | X-ray | 180 A | P | 39-51 | PDB |
| 7O5X | X-ray | 180 A | P | 39-51 | PDB |
| 7O6F | X-ray | 200 A | P | 39-51 | PDB |
| 7O6G | X-ray | 180 A | P | 39-51 | PDB |
| 7O6I | X-ray | 180 A | P | 39-51 | PDB |
| 7O6J | X-ray | 140 A | P | 39-51 | PDB |
| 7O6K | X-ray | 140 A | P | 39-51 | PDB |
| 7O6M | X-ray | 140 A | P | 39-51 | PDB |
| 7O6O | X-ray | 140 A | P | 39-51 | PDB |
| AF-Q04206-F1 | Predicted | AlphaFoldDB |
358 variants for Q04206
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA279852 rs863223356 RCV000202330 |
110 | I>T | Childhood-Onset Schizophrenia [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs750044010 CA6107009 |
4 | L>V | No |
ClinGen ExAC TOPMed |
|
|
rs1402366956 CA381395187 |
6 | P>S | No |
ClinGen gnomAD |
|
|
CA381395170 rs1169785805 |
8 | I>M | No |
ClinGen gnomAD |
|
|
rs1462514055 CA381395167 |
9 | F>V | No |
ClinGen gnomAD |
|
|
rs1263528129 CA381395161 |
10 | P>A | No |
ClinGen TOPMed |
|
|
rs368784411 CA224011379 |
10 | P>L | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA381395159 rs1263528129 |
10 | P>S | No |
ClinGen TOPMed |
|
|
rs1338619714 CA381395131 |
12 | E>D | No |
ClinGen gnomAD |
|
|
CA381395119 rs1332337161 |
14 | A>V | No |
ClinGen gnomAD |
|
|
rs1376292952 CA381395105 |
16 | A>V | No |
ClinGen gnomAD |
|
|
rs1418277571 CA381395104 |
17 | S>P | No |
ClinGen gnomAD |
|
|
CA381395087 rs1472798377 |
19 | P>L | No |
ClinGen gnomAD |
|
|
CA381395090 rs1159476176 |
19 | P>S | No |
ClinGen gnomAD |
|
|
CA6106985 rs765199827 |
20 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1213950702 CA381395033 |
27 | P>S | No |
ClinGen gnomAD |
|
|
rs753627793 CA6106983 |
28 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1285811443 CA381394973 |
36 | Y>H | No |
ClinGen gnomAD |
|
|
rs1268667021 CA381394949 |
39 | E>Q | No |
ClinGen TOPMed |
|
|
rs1193259548 CA381394925 |
42 | S>F | No |
ClinGen gnomAD |
|
|
CA6106979 rs764030082 |
45 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs969818412 CA224011265 |
49 | E>K | No |
ClinGen gnomAD |
|
|
CA381394877 rs1167688221 |
50 | R>K | No |
ClinGen gnomAD |
|
|
rs770416604 CA6106977 |
51 | S>R | No |
ClinGen ExAC gnomAD |
|
|
rs1565192440 COSM1744255 CA381394616 |
64 | N>S | biliary_tract [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA6106949 rs368661961 |
65 | G>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6106947 rs779944761 |
69 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381394549 rs1412466213 |
72 | V>L | No |
ClinGen gnomAD |
|
|
CA224011109 rs151125290 |
73 | R>C | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1438780274 CA381394539 |
73 | R>H | No |
ClinGen gnomAD |
|
|
CA381394487 rs1178830994 |
78 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1480907469 CA381394478 |
79 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA6106944 rs761767619 |
80 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381394452 rs1282651820 |
81 | P>R | No |
ClinGen gnomAD |
|
|
rs1484848625 CA381394456 |
81 | P>S | No |
ClinGen gnomAD |
|
|
CA381394432 rs1353372476 |
83 | H>Q | No |
ClinGen gnomAD |
|
|
rs752869747 CA6106942 |
84 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1590939582 CA381394409 |
86 | H>P | No |
ClinGen Ensembl |
|
|
CA224011083 rs1012952762 |
91 | V>L | No |
ClinGen TOPMed |
|
|
rs1374686489 CA381394342 |
93 | K>R | No |
ClinGen gnomAD |
|
|
rs11557248 CA224011078 |
96 | R>L | No |
ClinGen gnomAD |
|
|
rs11557248 CA381394307 |
96 | R>Q | No |
ClinGen gnomAD |
|
|
CA6106940 rs759715841 |
96 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766963304 CA6106938 |
97 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs562262364 CA6106937 |
100 | Y>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA381394270 rs1358445187 |
100 | Y>H | No |
ClinGen gnomAD |
|
|
CA6106936 rs773613820 |
104 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA381394226 rs773613820 |
104 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA6106933 rs774014347 |
106 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6106934 rs774014347 |
106 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774014347 CA381394202 |
106 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748761373 CA381394188 |
108 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1488068046 CA381394185 |
108 | R>H | No |
ClinGen gnomAD |
|
|
rs748761373 CA6106931 |
108 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA6106930 rs779601305 |
110 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769755404 CA6106929 |
111 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749535863 CA6106904 |
123 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA6106903 rs780140984 |
124 | R>Q | No |
ClinGen ExAC |
|
|
CA381393578 rs1172744425 |
127 | E>K | No |
ClinGen gnomAD |
|
|
rs1429314714 CA381393532 |
130 | I>V | No |
ClinGen gnomAD |
|
|
rs750564559 CA6106901 |
133 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6106900 rs767880564 |
135 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs757845338 CA6106899 |
136 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA381393403 rs1254116157 |
138 | N>I | No |
ClinGen gnomAD |
|
|
rs751905199 CA6106898 |
139 | N>T | No |
ClinGen ExAC gnomAD |
|
|
rs1336819380 CA381393365 |
141 | F>S | No |
ClinGen gnomAD |
|
|
rs1436789790 CA381393265 |
144 | P>H | No |
ClinGen gnomAD |
|
|
rs1158346001 CA381393246 |
145 | I>T | No |
ClinGen gnomAD |
|
|
rs140130811 CA224010176 |
145 | I>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1255403640 CA381393228 |
146 | E>G | No |
ClinGen TOPMed |
|
|
rs1469104216 CA381393210 |
147 | E>G | No |
ClinGen gnomAD |
|
|
rs1590937041 CA381393202 |
148 | Q>E | No |
ClinGen Ensembl |
|
|
CA381393192 rs1345639265 |
148 | Q>R | No |
ClinGen TOPMed |
|
|
rs1365998197 CA381393178 |
149 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
CA6106876 rs761250712 |
149 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381393172 rs761250712 |
149 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381393085 rs1398971779 |
157 | V>M | No |
ClinGen gnomAD |
|
|
rs375925395 CA6106871 |
158 | R>Q | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1457182302 CA381393010 |
164 | T>S | No |
ClinGen gnomAD |
|
|
CA224010160 rs957917859 |
165 | V>A | No |
ClinGen TOPMed |
|
|
CA6106868 rs774268451 |
166 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs568782744 CA6106869 COSM3810080 |
166 | R>W | breast [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC gnomAD |
|
CA381392942 rs1590936944 |
167 | D>A | No |
ClinGen Ensembl |
|
|
CA6106867 rs769890889 |
167 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA381392932 rs1416924926 |
168 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA381392924 rs1422039715 |
168 | P>L | No |
ClinGen TOPMed |
|
|
rs2375676 CA224010138 |
169 | S>A | No |
ClinGen Ensembl |
|
|
CA224010139 rs2375676 |
169 | S>T | No |
ClinGen Ensembl |
|
|
rs746111744 CA6106866 |
171 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746111744 CA381392875 |
171 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1399250604 CA381392872 |
172 | P>S | No |
ClinGen gnomAD |
|
|
CA381392850 rs1590936874 |
173 | L>P | No |
ClinGen Ensembl |
|
|
CA381392839 COSM1561661 rs1159602684 |
174 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1461359004 CA381392838 |
174 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA6106863 rs747469170 |
176 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1301308246 CA381392803 |
176 | P>S | No |
ClinGen TOPMed |
|
|
CA224010113 rs548164575 |
178 | V>L | No |
ClinGen Ensembl |
|
|
CA381392713 rs1475365161 |
181 | H>R | No |
ClinGen gnomAD |
|
|
rs1241103660 CA381392690 |
183 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA381391992 rs1393793185 |
189 | P>A | No |
ClinGen TOPMed |
|
|
rs1308599130 CA381391945 |
193 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1308599130 CA381391943 |
193 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA224009282 rs11606329 |
197 | C>R | No |
ClinGen Ensembl |
|
|
RCV001092067 rs1326268854 |
198 | R>* | No |
ClinVar dbSNP |
|
|
CA6106838 rs145138143 |
199 | V>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs755895938 CA6106836 |
201 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1194237734 CA381391748 |
207 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA381391631 rs1439792199 |
216 | C>G | No |
ClinGen gnomAD |
|
|
rs1439805810 CA381391453 |
226 | V>M | No |
ClinGen TOPMed |
|
|
CA381391436 rs1314465477 |
227 | Y>C | No |
ClinGen TOPMed |
|
|
CA6106817 rs368076224 |
228 | F>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs756811835 CA6106816 |
229 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1032578811 CA224009025 |
230 | G>R | No |
ClinGen TOPMed |
|
|
rs764063700 CA6106814 |
231 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs751077868 CA6106815 |
231 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1343398488 CA381391366 |
232 | G>A | No |
ClinGen gnomAD |
|
|
CA381391325 rs1363382539 |
234 | E>G | No |
ClinGen TOPMed |
|
|
rs1856484601 RCV001055525 |
236 | R>* | No |
ClinVar dbSNP |
|
|
CA6106811 rs765033710 |
236 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA224008981 rs926385426 |
242 | A>G | No |
ClinGen Ensembl |
|
|
rs1474354942 CA381391151 |
244 | V>G | No |
ClinGen gnomAD |
|
|
CA381391120 RCV000754618 rs1565190345 |
246 | R>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA6106806 rs774359103 |
250 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA6106805 rs769190277 |
253 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs769190277 CA381390994 |
253 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1590935322 CA381390988 |
254 | T>P | No |
ClinGen Ensembl |
|
|
CA6106802 rs770313905 |
258 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745468574 CA381390914 |
259 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA6106801 rs745468574 |
259 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA6106800 rs780968051 |
260 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA381390888 rs1381464457 |
261 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
CA381390887 rs1381464457 |
261 | S>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1359669742 CA381390871 |
263 | Q>P | No |
ClinGen gnomAD |
|
|
CA6106799 rs770531864 |
264 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs746526485 CA6106798 |
264 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381390851 rs1393561951 |
265 | P>A | No |
ClinGen gnomAD |
|
|
CA6106796 rs758298850 |
266 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758298850 CA6106797 |
266 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381390828 rs1420318686 COSM1223509 |
267 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
rs1420318686 CA381390830 |
267 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA381390827 rs1165809846 |
267 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs866716163 CA224008912 |
269 | S>F | No |
ClinGen Ensembl |
|
|
CA6106794 rs778642588 |
273 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381390754 rs1417895912 |
273 | R>W | No |
ClinGen gnomAD |
|
|
CA13419741 rs1252847592 |
274 | R>W | No |
ClinGen gnomAD |
|
|
rs149254469 CA6106789 |
278 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6106790 rs761995751 |
278 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs763514130 CA6106788 |
280 | L>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6106787 rs763514130 |
280 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs776162117 CA6106786 |
281 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1370194169 CA381390665 |
281 | S>T | No |
ClinGen gnomAD |
|
|
CA381390603 rs1323750358 |
285 | E>D | No |
ClinGen gnomAD |
|
|
rs929817187 CA224008883 |
285 | E>K | No |
ClinGen TOPMed |
|
|
CA381390528 rs1237054467 |
291 | D>A | No |
ClinGen TOPMed |
|
|
rs61759893 CA6106785 |
291 | D>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6106784 rs759945679 |
292 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1172994773 CA381390513 |
293 | D>N | No |
ClinGen gnomAD |
|
|
rs374605673 CA381389858 |
294 | D>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374605673 CA6106756 |
294 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6106755 rs749182844 |
295 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA6106754 rs150601443 |
295 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA381389832 rs1565189032 |
296 | H>L | No |
ClinGen Ensembl |
|
|
CA6106752 COSM930488 rs142033286 |
297 | R>Q | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs755884402 CA6106753 |
297 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs377717312 CA224006623 |
301 | K>T | No |
ClinGen ESP |
|
|
CA224006622 rs368638080 |
302 | R>C | No |
ClinGen Ensembl |
|
|
CA381389744 rs1565188999 |
303 | K>* | No |
ClinGen Ensembl |
|
|
rs1254886027 CA381389731 |
304 | R>K | No |
ClinGen gnomAD |
|
|
rs138034120 CA6106748 |
306 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA381389676 rs1260736909 |
308 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
rs755476416 CA6106747 |
312 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA6106746 rs12721572 |
313 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1191351823 CA381389610 |
314 | K>E | No |
ClinGen TOPMed |
|
|
CA381389590 rs1334453316 |
316 | S>G | No |
ClinGen gnomAD |
|
|
rs1291631162 CA381389577 |
317 | P>T | No |
ClinGen gnomAD |
|
|
CA6106744 rs760929899 |
318 | F>S | No |
ClinGen ExAC gnomAD |
|
|
CA381389551 rs1466629537 |
319 | S>N | No |
ClinGen TOPMed |
|
|
rs767155175 CA6106742 |
320 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381389541 rs767155175 |
320 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381389469 rs1196982543 |
321 | P>L | No |
ClinGen gnomAD |
|
|
rs999556978 CA224006460 |
321 | P>T | No |
ClinGen Ensembl |
|
|
CA6106716 rs770882250 |
322 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA6106715 rs375929528 |
322 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs749325969 CA6106712 |
323 | D>E | No |
ClinGen ExAC gnomAD |
|
|
rs138794116 CA6106713 |
323 | D>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs780241287 CA6106711 |
324 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs762060621 CA381389408 |
325 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762060621 CA6106710 |
325 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1161370675 CA381389414 |
325 | R>W | No |
ClinGen TOPMed |
|
|
rs750957760 CA6106709 |
326 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381389406 rs750957760 |
326 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381389377 rs1432792806 |
327 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA381389343 rs1193014025 |
329 | R>G | No |
ClinGen gnomAD |
|
|
rs1282218073 CA381389336 |
329 | R>Q | No |
ClinGen TOPMed |
|
|
COSM1223508 CA381389330 rs1478374154 |
330 | R>C | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1232341468 CA381389322 COSM930486 |
330 | R>H | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1180352560 CA381389311 |
331 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA381389256 rs1484150455 |
334 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA6106707 rs757592217 |
335 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA381389234 rs757592217 |
335 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs751960651 CA6106706 |
336 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1246637051 CA381389221 |
336 | R>H | No |
ClinGen gnomAD |
|
|
CA381389212 rs1384344058 |
337 | S>C | No |
ClinGen TOPMed gnomAD |
|
|
rs1384344058 CA381389214 |
337 | S>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1219333291 CA381389208 |
337 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1219333291 CA381389211 |
337 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs762722479 CA6106704 |
339 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1411351659 CA381389157 |
340 | S>P | No |
ClinGen gnomAD |
|
|
CA381389135 rs1312067163 |
341 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs371744052 CA6106701 |
343 | K>N | No |
ClinGen ESP ExAC gnomAD |
|
|
CA6106702 rs764810980 |
343 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA6106699 rs771045696 |
344 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA224005990 rs997637338 |
347 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA381388428 rs1196154107 |
348 | P>S | No |
ClinGen gnomAD |
|
|
rs1267397579 CA381388399 |
350 | P>T | No |
ClinGen gnomAD |
|
|
rs1481868107 CA381388369 |
352 | T>A | No |
ClinGen gnomAD |
|
|
CA6106650 rs774079396 |
352 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381388329 rs1347094026 |
354 | S>F | No |
ClinGen gnomAD |
|
|
rs1235283948 CA381388311 |
356 | S>G | No |
ClinGen gnomAD |
|
|
rs759766423 CA6106648 |
357 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs1302055993 CA381388274 |
360 | Y>C | No |
ClinGen gnomAD |
|
|
rs1310254235 CA381388252 |
361 | D>E | No |
ClinGen gnomAD |
|
|
rs1348785979 CA381388266 |
361 | D>N | No |
ClinGen gnomAD |
|
|
rs1410768522 CA381388212 |
364 | P>L | No |
ClinGen gnomAD |
|
|
rs1402154056 CA381388205 |
365 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA381388184 rs1171374836 |
366 | M>T | No |
ClinGen gnomAD |
|
|
rs12721575 CA224005933 |
368 | F>L | No |
ClinGen Ensembl |
|
|
rs12721575 CA224005931 |
368 | F>L | No |
ClinGen Ensembl |
|
|
rs1468006702 CA381388140 |
369 | P>L | No |
ClinGen gnomAD |
|
|
rs771032052 CA6106646 |
370 | S>A | No |
ClinGen ExAC gnomAD |
|
|
CA6106645 rs747467496 |
371 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381388124 rs1157690704 |
371 | G>R | No |
ClinGen gnomAD |
|
|
CA6106644 rs773748330 |
373 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs772497103 CA6106643 |
375 | Q>E | No |
ClinGen ExAC |
|
|
rs1485245702 CA381388049 |
376 | A>D | No |
ClinGen gnomAD |
|
|
rs748545913 CA6106642 |
377 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs1483533790 CA381388033 |
378 | A>T | No |
ClinGen gnomAD |
|
|
CA6106640 rs754622774 |
380 | A>D | No |
ClinGen ExAC |
|
|
rs1290399583 CA381387994 |
381 | P>L | No |
ClinGen gnomAD |
|
|
CA381387957 rs1332591254 |
384 | P>R | No |
ClinGen gnomAD |
|
|
rs1405172010 CA381387961 |
384 | P>S | No |
ClinGen TOPMed |
|
|
rs755660407 CA6106637 |
385 | Q>H | No |
ClinGen ExAC gnomAD |
|
|
rs779659272 CA6106638 |
385 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1413493308 CA381387901 |
388 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1283746730 CA381387907 |
388 | P>S | No |
ClinGen gnomAD |
|
|
rs1366570652 CA381387878 |
390 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1346474543 CA381387872 |
391 | P>S | No |
ClinGen TOPMed |
|
|
rs1189280546 CA381387866 |
392 | A>T | No |
ClinGen gnomAD |
|
|
CA381387852 rs1260899484 |
393 | P>S | No |
ClinGen gnomAD |
|
|
rs1208220510 CA381387835 |
395 | P>R | No |
ClinGen gnomAD |
|
|
rs1463629801 CA381387830 |
396 | A>T | No |
ClinGen gnomAD |
|
|
CA381387811 rs1209657498 |
397 | P>L | No |
ClinGen gnomAD |
|
|
rs749937116 CA6106636 |
397 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1355903094 CA381387808 |
398 | A>S | No |
ClinGen gnomAD |
|
|
CA381387803 rs1282808983 |
398 | A>V | No |
ClinGen gnomAD |
|
|
CA381387791 rs1341628692 |
399 | M>I | No |
ClinGen gnomAD |
|
|
rs560604471 CA381387782 |
400 | V>I | No |
ClinGen gnomAD |
|
|
rs560604471 CA224005827 |
400 | V>L | No |
ClinGen gnomAD |
|
|
CA381387752 rs1263055359 |
403 | L>V | No |
ClinGen TOPMed |
|
|
rs1390278423 CA381387743 |
404 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
CA381387744 rs1390278423 |
404 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs781182014 CA6106634 |
406 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1590931358 CA381387696 |
407 | P>L | No |
ClinGen Ensembl |
|
|
CA6106632 rs751310615 |
408 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA381387677 rs1210908822 |
409 | P>L | No |
ClinGen TOPMed |
|
|
CA381387611 rs1451237617 |
415 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
CA381387620 rs1186645003 |
415 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA6106628 rs754083713 |
418 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs375768034 CA381387551 |
420 | A>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA6106626 rs375768034 |
420 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1287948980 CA381387546 |
421 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1012204019 CA224005761 |
422 | A>V | No |
ClinGen TOPMed |
|
|
rs1211831291 CA381387505 |
426 | P>L | No |
ClinGen gnomAD |
|
|
rs773234750 CA6106624 |
427 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
CA381387481 rs1590931266 |
429 | T>A | No |
ClinGen Ensembl |
|
|
rs1297711254 CA381387464 |
430 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs55766259 CA224005732 |
431 | A>G | No |
ClinGen Ensembl |
|
|
rs774718207 CA6106622 |
433 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381387414 rs1329027216 |
434 | G>A | No |
ClinGen gnomAD |
|
|
CA6106621 rs555785944 |
435 | T>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6106619 rs749716796 |
437 | S>* | No |
ClinGen ExAC gnomAD |
|
|
CA381387370 rs1431554156 |
438 | E>G | No |
ClinGen gnomAD |
|
|
CA381387356 rs1390663138 |
439 | A>D | No |
ClinGen gnomAD |
|
|
rs1390663138 CA381387354 |
439 | A>V | No |
ClinGen gnomAD |
|
|
CA381387333 rs1168603207 |
442 | Q>* | No |
ClinGen gnomAD |
|
|
rs11608099 CA224005727 |
442 | Q>H | No |
ClinGen Ensembl |
|
|
CA6106617 rs1477765782 |
442 | Q>L | No |
ClinGen TOPMed gnomAD |
|
|
CA381387330 rs1477765782 |
442 | Q>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1490573963 CA381387309 |
444 | Q>R | No |
ClinGen gnomAD |
|
|
rs1485728798 CA381387277 |
446 | D>E | No |
ClinGen TOPMed |
|
|
CA381387278 rs1430719920 |
446 | D>V | No |
ClinGen gnomAD |
|
|
rs769633553 CA381387263 |
447 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs780580304 CA6106613 |
449 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381387246 rs1344961447 |
449 | D>H | No |
ClinGen gnomAD |
|
|
rs1219124903 CA381387207 |
452 | A>V | No |
ClinGen gnomAD |
|
|
rs757149780 CA6106611 |
455 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs758168856 CA6106608 |
458 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758168856 CA6106609 |
458 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs367873849 CA6106607 |
460 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1411388720 CA381387088 |
462 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA381387067 rs1333488473 |
463 | F>L | No |
ClinGen gnomAD |
|
|
rs766584902 CA6106606 |
463 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
CA6106605 rs760866901 |
464 | T>A | No |
ClinGen ExAC gnomAD |
|
|
CA381387024 rs1174324125 |
467 | A>V | No |
ClinGen gnomAD |
|
|
CA6106600 rs769322382 |
470 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381386972 rs1366985822 |
471 | N>S | No |
ClinGen TOPMed |
|
|
CA6106598 rs535576789 |
473 | E>D | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1177462012 CA381386950 |
473 | E>K | No |
ClinGen gnomAD |
|
|
rs769429371 CA6106597 |
475 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs1393696800 CA381386915 |
476 | Q>H | No |
ClinGen TOPMed |
|
|
CA6106596 rs745510018 |
481 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381386874 rs1267463921 |
482 | I>V | No |
ClinGen TOPMed |
|
|
CA6106595 rs374751242 |
485 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6106594 rs566890727 |
486 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6106592 rs777658303 |
487 | H>P | No |
ClinGen ExAC gnomAD |
|
|
rs546654982 CA224005622 |
487 | H>Q | No |
ClinGen 1000Genomes gnomAD |
|
|
rs1283377121 CA381386754 |
492 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
rs747928226 CA6106590 |
493 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA6106588 rs756297857 |
495 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs778695678 CA6106589 |
495 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1387957052 CA381386666 |
499 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA381386660 rs1157278470 |
500 | I>V | No |
ClinGen gnomAD |
|
|
CA6106586 rs767826580 |
502 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1181568528 CA381386640 |
502 | R>H | No |
ClinGen gnomAD |
|
|
rs1239685921 CA381386613 |
505 | T>S | No |
ClinGen gnomAD |
|
|
CA381386606 rs1565187728 |
506 | G>A | No |
ClinGen Ensembl |
|
|
CA6106584 rs751753627 |
506 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs540158560 CA6106583 |
507 | A>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1363246020 CA381386585 |
509 | R>S | No |
ClinGen TOPMed |
|
|
CA381386570 rs1463059248 |
512 | D>N | No |
ClinGen TOPMed |
|
|
rs1463059248 CA381386571 |
512 | D>Y | No |
ClinGen TOPMed |
|
|
CA381386559 rs1381286744 |
513 | P>L | No |
ClinGen gnomAD |
|
|
CA381386561 rs1381286744 |
513 | P>Q | No |
ClinGen gnomAD |
|
|
rs200767506 CA381386562 |
513 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs200767506 CA6106580 |
513 | P>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs369008511 CA224005573 |
514 | A>G | No |
ClinGen ESP TOPMed |
|
|
rs1590930882 CA381386539 |
517 | P>T | No |
ClinGen Ensembl |
|
|
rs1364620549 CA381386516 |
519 | G>A | No |
ClinGen gnomAD |
|
|
CA6106577 rs776126630 |
520 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1345948863 CA381386493 |
521 | P>L | No |
ClinGen gnomAD |
|
|
rs746519095 CA6106575 |
525 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6106574 rs201037601 |
528 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6106573 rs201037601 |
528 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1184817070 CA381386411 |
529 | S>P | No |
ClinGen gnomAD |
|
|
rs778642669 CA6106571 |
530 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs754755213 CA6106570 |
532 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA381386352 rs1189873448 |
532 | E>G | No |
ClinGen gnomAD |
|
|
CA224005537 rs767319473 |
533 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA381386323 rs767319473 |
533 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1382122315 CA381386274 |
537 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs199846198 CA6106568 |
538 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs754554427 CA224005517 |
540 | M>T | No |
ClinGen Ensembl |
|
|
rs375525158 CA6106565 |
540 | M>V | No |
ClinGen ESP ExAC gnomAD |
|
|
CA381386156 rs1227655115 |
542 | F>L | No |
ClinGen gnomAD |
|
|
rs758932077 CA6106563 |
544 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA224005501 rs372036028 |
546 | L>M | No |
ClinGen ESP |
|
|
rs951877648 CA224005490 |
548 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs759788260 CA6106560 |
550 | S>T | No |
ClinGen ExAC gnomAD |
2 associated diseases with Q04206
[MIM: 618287]: Mucocutaneous ulceration, chronic (CMCU)
An autosomal dominant, mucocutaneous disease characterized by chronic mucosal lesions, in absence of recurrent infections. {ECO:0000269|PubMed:28600438}. Note=The disease may be caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant, mucocutaneous disease characterized by chronic mucosal lesions, in absence of recurrent infections. {ECO:0000269|PubMed:28600438}. Note=The disease may be caused by variants affecting the gene represented in this entry.
6 regional properties for Q04206
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | IPT domain | 193 - 289 | IPR002909 |
| domain | Rel homology domain, DNA-binding domain | 16 - 190 | IPR011539 |
| conserved_site | Rel homology domain, conserved site | 34 - 40 | IPR030492 |
| domain | Transcription factor p65, RHD domain, N-terminal | 19 - 187 | IPR030495 |
| domain | Rel homology dimerisation domain | 195 - 291 | IPR032397 |
| domain | NFkappaB IPT domain | 194 - 290 | IPR033926 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24169 | NUCLEAR FACTOR NF-KAPPA-B PROTEIN |
| PANTHER Subfamily | PTHR24169:SF1 | TRANSCRIPTION FACTOR P65 |
| PANTHER Protein Class |
DNA-binding transcription factor
Rel homology transcription factor immunoglobulin fold transcription factor |
|
| PANTHER Pathway Category |
Inflammation mediated by chemokine and cytokine signaling pathway NFkappaB Gonadotropin-releasing hormone receptor pathway p65 Toll receptor signaling pathway NFkappaB Apoptosis signaling pathway NFkappaB |
|
9 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| NF-kappaB complex | A protein complex that consists of a homo- or heterodimer of members of a family of structurally related proteins that contain a conserved N-terminal region called the Rel homology domain (RHD). In the nucleus, NF-kappaB complexes act as transcription factors. In unstimulated cells, NF-kappaB dimers are sequestered in the cytoplasm by IkappaB monomers; signals that induce NF-kappaB activity cause degradation of IkappaB, allowing NF-kappaB dimers to translocate to the nucleus and induce gene expression. |
| NF-kappaB p50/p65 complex | A heterodimer of NF-kappa B p50 and p65 subunits. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| transcription regulator complex | A protein complex that is capable of associating with DNA by direct binding, or via other DNA-binding proteins or complexes, and regulating transcription. |
26 GO annotations of molecular function
| Name | Definition |
|---|---|
| actinin binding | Binding to actinin, any member of a family of proteins that crosslink F-actin. |
| ankyrin repeat binding | Binding to an ankyrin repeat of a protein. Ankyrin repeats are tandemly repeated modules of about 33 amino acids; each repeat folds into a helix-loop-helix structure with a beta-hairpin/loop region projecting out from the helices at a 90-degree angle, and repeats stack to form an L-shaped structure. |
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| chromatin DNA binding | Binding to DNA that is assembled into chromatin. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| DNA-binding transcription repressor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that represses or decreases the transcription of specific gene sets transcribed by RNA polymerase II. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| general transcription initiation factor binding | Binding to a general transcription initiation factor, a protein that contributes to transcription start site selection and transcription initiation. |
| histone deacetylase binding | Binding to histone deacetylase. |
| identical protein binding | Binding to an identical protein or proteins. |
| NF-kappaB binding | Binding to NF-kappaB, a transcription factor for eukaryotic RNA polymerase II promoters. |
| peptide binding | Binding to a peptide, an organic compound comprising two or more amino acids linked by peptide bonds. |
| phosphate ion binding | Binding to a phosphate ion. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein N-terminus binding | Binding to a protein N-terminus, the end of any peptide chain at which the 2-amino (or 2-imino) function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II core promoter sequence-specific DNA binding | Binding to a DNA sequence that is part of the core promoter of a RNA polymerase II-transcribed gene. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| transcription cis-regulatory region binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls transcription of that section of the DNA. The transcribed region might be described as a gene, cistron, or operon. |
| transcription coactivator binding | Binding to a transcription coactivator, a protein involved in positive regulation of transcription via protein-protein interactions with transcription factors and other proteins that positively regulate transcription. Transcription coactivators do not bind DNA directly, but rather mediate protein-protein interactions between activating transcription factors and the basal transcription machinery. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
62 GO annotations of biological process
| Name | Definition |
|---|---|
| animal organ morphogenesis | Morphogenesis of an animal organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process in which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| cellular defense response | A defense response that is mediated by cells. |
| cellular response to angiotensin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an angiotensin stimulus. Angiotensin is any of three physiologically active peptides (angiotensin II, III, or IV) processed from angiotensinogen. |
| cellular response to hepatocyte growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hepatocyte growth factor stimulus. |
| cellular response to hydrogen peroxide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| cellular response to interleukin-1 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| cellular response to interleukin-6 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-6 stimulus. |
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| cellular response to lipoteichoic acid | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipoteichoic acid stimulus; lipoteichoic acid is a major component of the cell wall of gram-positive bacteria and typically consists of a chain of glycerol-phosphate repeating units linked to a glycolipid anchor. |
| cellular response to nicotine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a nicotine stimulus. |
| cellular response to peptidoglycan | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptidoglycan stimulus. Peptidoglycan is a bacterial cell wall macromolecule. |
| cellular response to tumor necrosis factor | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a tumor necrosis factor stimulus. |
| cellular response to vascular endothelial growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a vascular endothelial growth factor stimulus. |
| chromatin organization | The assembly or remodeling of chromatin composed of DNA complexed with histones, other associated proteins, and sometimes RNA. |
| cytokine-mediated signaling pathway | The series of molecular signals initiated by the binding of a cytokine to a receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| defense response to virus | Reactions triggered in response to the presence of a virus that act to protect the cell or organism. |
| hair follicle development | The process whose specific outcome is the progression of the hair follicle over time, from its formation to the mature structure. A hair follicle is a tube-like opening in the epidermis where the hair shaft develops and into which the sebaceous glands open. |
| I-kappaB kinase/NF-kappaB signaling | The process in which a signal is passed on to downstream components within the cell through the I-kappaB-kinase (IKK)-dependent activation of NF-kappaB. The cascade begins with activation of a trimeric IKK complex (consisting of catalytic kinase subunits IKKalpha and/or IKKbeta, and the regulatory scaffold protein NEMO) and ends with the regulation of transcription of target genes by NF-kappaB. In a resting state, NF-kappaB dimers are bound to I-kappaB proteins, sequestering NF-kappaB in the cytoplasm. Phosphorylation of I-kappaB targets I-kappaB for ubiquitination and proteasomal degradation, thus releasing the NF-kappaB dimers, which can translocate to the nucleus to bind DNA and regulate transcription. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| interleukin-1-mediated signaling pathway | The series of molecular signals initiated by interleukin-1 binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| liver development | The process whose specific outcome is the progression of the liver over time, from its formation to the mature structure. The liver is an exocrine gland which secretes bile and functions in metabolism of protein and carbohydrate and fat, synthesizes substances involved in the clotting of the blood, synthesizes vitamin A, detoxifies poisonous substances, stores glycogen, and breaks down worn-out erythrocytes. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of extrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway. |
| negative regulation of NIK/NF-kappaB signaling | Any process that stops, prevents or reduces the frequency, rate or extent of NIK/NF-kappaB signaling. |
| negative regulation of pri-miRNA transcription by RNA polymerase II | Any process that stops, prevents or reduces the frequency, rate or extent of microRNA (miRNA) gene transcription. |
| negative regulation of protein catabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of protein catabolic process. |
| negative regulation of protein sumoylation | Any process that stops, prevents, or reduces the frequency, rate or extent of the addition of SUMO groups to a protein. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| negative regulation of transcription, DNA-templated | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| neuropeptide signaling pathway | A G protein-coupled receptor signaling pathway initiated by a neuropeptide binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process. |
| NIK/NF-kappaB signaling | The process in which a signal is passed on to downstream components within the cell through the NIK-dependent processing and activation of NF-KappaB. Begins with activation of the NF-KappaB-inducing kinase (NIK), which in turn phosphorylates and activates IkappaB kinase alpha (IKKalpha). IKKalpha phosphorylates the NF-Kappa B2 protein (p100) leading to p100 processing and release of an active NF-KappaB (p52). |
| nucleotide-binding oligomerization domain containing 2 signaling pathway | The series of molecular signals initiated by the binding of a ligand (such as a bacterial peptidoglycan) to a cytoplasmic nucleotide-binding oligomerization domain containing 2 (NOD2) protein receptor, and ending with regulation of a downstream cellular process. |
| positive regulation of amyloid-beta formation | Any process that activates or increases the frequency, rate or extent of amyloid-beta formation. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of I-kappaB kinase/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of interleukin-12 production | Any process that activates or increases the frequency, rate, or extent of interleukin-12 production. |
| positive regulation of interleukin-8 production | Any process that activates or increases the frequency, rate, or extent of interleukin-8 production. |
| positive regulation of leukocyte adhesion to vascular endothelial cell | Any process that activates or increases the frequency, rate or extent of leukocyte adhesion to vascular endothelial cell. |
| positive regulation of miRNA metabolic process | Any process that activates or increases the frequency, rate or extent of miRNA metabolic process. |
| positive regulation of NF-kappaB transcription factor activity | Any process that activates or increases the frequency, rate or extent of activity of the transcription factor NF-kappaB. |
| positive regulation of NIK/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of NIK/NF-kappaB signaling. |
| positive regulation of pri-miRNA transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of microRNA (miRNA) gene transcription. |
| positive regulation of T cell receptor signaling pathway | Any process that activates or increases the frequency, rate or extent of signaling pathways initiated by the cross-linking of an antigen receptor on a T cell. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus | Any positive regulation of transcription from RNA polymerase II promoter that is involved in cellular response to chemical stimulus. |
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| postsynapse to nucleus signaling pathway | The series of molecular signals that conveys information from the postsynapse to the nucleus via cytoskeletal transport of a protein from a postsynapse to the component to the nucleus where it affects biochemical processes that occur in the nucleus (e.g DNA transcription, mRNA splicing, or DNA/histone modifications). |
| regulation of DNA-templated transcription in response to stress | Modulation of the frequency, rate or extent of transcription from a DNA template as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation). |
| regulation of inflammatory response | Any process that modulates the frequency, rate or extent of the inflammatory response, the immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. |
| regulation of NIK/NF-kappaB signaling | Any process that modulates the frequency, rate or extent of NIK/NF-kappaB signaling. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| regulation of transcription, DNA-templated | Any process that modulates the frequency, rate or extent of cellular DNA-templated transcription. |
| response to cytokine | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a cytokine stimulus. |
| response to interleukin-1 | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| response to muramyl dipeptide | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a muramyl dipeptide stimulus. Muramyl dipeptide is derived from peptidoglycan. |
| response to muscle stretch | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a myofibril being extended beyond its slack length. |
| response to organic substance | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an organic substance stimulus. |
| response to UV-B | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a UV-B radiation stimulus. UV-B radiation (UV-B light) spans the wavelengths 280 to 315 nm. |
| transcription, DNA-templated | The synthesis of an RNA transcript from a DNA template. |
| tumor necrosis factor-mediated signaling pathway | The series of molecular signals initiated by tumor necrosis factor binding to its receptor on the surface of a cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q94527 | Rel | Nuclear factor NF-kappa-B p110 subunit | Drosophila melanogaster (Fruit fly) | EV |
| Q00653 | NFKB2 | Nuclear factor NF-kappa-B p100 subunit | Homo sapiens (Human) | SS |
| P19838 | NFKB1 | Nuclear factor NF-kappa-B p105 subunit | Homo sapiens (Human) | SS |
| Q04207 | Rela | Transcription factor p65 | Mus musculus (Mouse) | SS |
| Q9WTK5 | Nfkb2 | Nuclear factor NF-kappa-B p100 subunit [Cleaved into: Nuclear factor NF-kappa-B p52 subunit] | Mus musculus (Mouse) | EV |
| P25799 | Nfkb1 | Nuclear factor NF-kappa-B p105 subunit [Cleaved into: Nuclear factor NF-kappa-B p50 subunit] | Mus musculus (Mouse) | EV |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDELFPLIFP | AEPAQASGPY | VEIIEQPKQR | GMRFRYKCEG | RSAGSIPGER | STDTTKTHPT |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IKINGYTGPG | TVRISLVTKD | PPHRPHPHEL | VGKDCRDGFY | EAELCPDRCI | HSFQNLGIQC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VKKRDLEQAI | SQRIQTNNNP | FQVPIEEQRG | DYDLNAVRLC | FQVTVRDPSG | RPLRLPPVLS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| HPIFDNRAPN | TAELKICRVN | RNSGSCLGGD | EIFLLCDKVQ | KEDIEVYFTG | PGWEARGSFS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QADVHRQVAI | VFRTPPYADP | SLQAPVRVSM | QLRRPSDREL | SEPMEFQYLP | DTDDRHRIEE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KRKRTYETFK | SIMKKSPFSG | PTDPRPPPRR | IAVPSRSSAS | VPKPAPQPYP | FTSSLSTINY |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DEFPTMVFPS | GQISQASALA | PAPPQVLPQA | PAPAPAPAMV | SALAQAPAPV | PVLAPGPPQA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VAPPAPKPTQ | AGEGTLSEAL | LQLQFDDEDL | GALLGNSTDP | AVFTDLASVD | NSEFQQLLNQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| GIPVAPHTTE | PMLMEYPEAI | TRLVTGAQRP | PDPAPAPLGA | PGLPNGLLSG | DEDFSSIADM |
| 550 | |||||
| DFSALLSQIS | S |