Q02078
Gene name |
MEF2A (MEF2) |
Protein name |
Myocyte-specific enhancer factor 2A |
Names |
Serum response factor-like protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4205 |
EC number |
|
Protein Class |
|
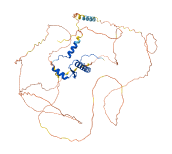
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1-61 (Transcription factor, MADS-box) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

9 structures for Q02078
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1C7U | NMR | - | A/B | 2-86 | PDB |
| 1EGW | X-ray | 150 A | A/B/C/D | 2-78 | PDB |
| 1LEW | X-ray | 230 A | B | 269-280 | PDB |
| 3KOV | X-ray | 290 A | A/B/I/J | 2-91 | PDB |
| 3MU6 | X-ray | 243 A | A/B/C/D | 2-70 | PDB |
| 3P57 | X-ray | 219 A | A/B/C/D/I/J | 2-91 | PDB |
| 6BYY | X-ray | 230 A | PDB | ||
| 6BZ1 | X-ray | 297 A | PDB | ||
| AF-Q02078-F1 | Predicted | AlphaFoldDB |
475 variants for Q02078
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV003390664 CA120017 rs121918530 RCV000420077 VAR_038407 RCV000009506 |
263 | N>S | Coronary artery disease/myocardial infarction [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_038408 RCV000009505 rs121918529 CA120015 |
279 | P>L | Coronary artery disease/myocardial infarction [ClinVar] | Yes |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
VAR_038409 rs121918531 RCV000009507 CA120019 |
283 | G>D | Coronary artery disease/myocardial infarction [ClinVar] | Yes |
ClinGen ClinVar UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
rs3138597 RCV003454973 RCV000959247 |
429 | Q>missing | Coronary artery disease, autosomal dominant, 1 [ClinVar] | Yes |
ClinVar dbSNP |
| rs1317833270 | 3 | R>Q | No | gnomAD | |
| rs1272954718 | 9 | T>R | No | gnomAD | |
| rs755726096 | 10 | R>C | No |
ExAC gnomAD |
|
| rs1206225537 | 10 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1206225537 | 10 | R>L | No |
TOPMed gnomAD |
|
| rs755726096 | 10 | R>S | No |
ExAC gnomAD |
|
| rs1376663731 | 12 | M>I | No | Ensembl | |
| rs766169740 | 12 | M>L | No |
ExAC gnomAD |
|
| rs868326714 | 14 | E>K | No | Ensembl | |
| rs753575051 | 15 | R>M | No |
ExAC gnomAD |
|
| rs1005984466 | 17 | R>* | No | Ensembl | |
| rs754720431 | 17 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs754720431 | 17 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2153479402 | 24 | R>G | No | Ensembl | |
| rs1350119833 | 28 | L>F | No | gnomAD | |
| rs2045835798 | 29 | M>I | No | Ensembl | |
| rs760337010 | 29 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs1291551374 | 33 | Y>C | No | TOPMed | |
| rs1434408562 | 35 | L>F | No | gnomAD | |
| rs2153479509 | 38 | L>P | No | 1000Genomes | |
| rs1328677553 | 40 | D>E | No | gnomAD | |
|
rs1226726746 RCV001354903 |
41 | C>* | No |
ClinVar dbSNP |
|
| rs1226726746 | 41 | C>* | No |
TOPMed gnomAD |
|
| rs2045840917 | 44 | A>V | No | TOPMed | |
| rs368747387 | 48 | F>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1226882901 | 49 | N>S | No |
TOPMed gnomAD |
|
| rs754843909 | 50 | S>T | No |
ExAC gnomAD |
|
| rs1343245101 | 52 | N>D | No | gnomAD | |
| rs2045844594 | 53 | K>R | No |
TOPMed gnomAD |
|
| rs764920921 | 55 | F>V | No |
ExAC gnomAD |
|
| rs1207528298 | 56 | Q>E | No | gnomAD | |
| rs2045848123 | 60 | T>I | No | TOPMed | |
| rs1193702038 | 61 | D>H | No |
TOPMed gnomAD |
|
| rs1193702038 | 61 | D>N | No |
TOPMed gnomAD |
|
| rs2045849231 | 61 | D>V | No | TOPMed | |
| rs781433971 | 62 | M>T | No |
ExAC gnomAD |
|
| rs746001869 | 63 | D>G | No |
ExAC gnomAD |
|
| rs1367123469 | 64 | K>T | No | Ensembl | |
| rs938477439 | 70 | T>R | No | TOPMed | |
| rs2153479814 | 72 | Y>H | No | Ensembl | |
|
COSM3667693 rs1456819815 |
73 | N>S | liver [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2045855834 | 74 | E>D | No |
TOPMed gnomAD |
|
| rs2045856958 | 78 | S>G | No | TOPMed | |
| rs1277082906 | 80 | T>N | No | gnomAD | |
| rs553844161 | 82 | S>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779104807 | 85 | V>A | No |
ExAC gnomAD |
|
| rs1167885533 | 87 | A>S | No | TOPMed | |
| rs1167885533 | 87 | A>T | No | TOPMed | |
| rs762881136 | 89 | N>K | No |
ExAC gnomAD |
|
| rs775203708 | 89 | N>S | No |
ExAC gnomAD |
|
| rs377723529 | 91 | K>R | No | ESP | |
| rs763955639 | 92 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs761799644 | 96 | C>Y | No |
ExAC TOPMed gnomAD |
|
| rs1184781770 | 97 | D>N | No |
TOPMed gnomAD |
|
| rs976656919 | 98 | S>T | No | TOPMed | |
| rs1374499801 | 99 | P>L | No |
TOPMed gnomAD |
|
| rs753805725 | 99 | P>S | No |
ExAC gnomAD |
|
| rs1462190719 | 101 | P>S | No | gnomAD | |
| rs1452466090 | 103 | T>I | No |
TOPMed gnomAD |
|
| rs1452466090 | 103 | T>S | No |
TOPMed gnomAD |
|
| rs755075913 | 104 | S>* | No |
ExAC gnomAD |
|
| rs201205600 | 105 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs1337363619 | 105 | Y>H | No | gnomAD | |
| rs2153630039 | 106 | V>G | No | Ensembl | |
| rs2153630047 | 108 | T>A | No | Ensembl | |
| rs2050876501 | 108 | T>I | No | TOPMed | |
| rs2050877041 | 109 | P>S | No | Ensembl | |
| rs1326741833 | 110 | H>L | No |
TOPMed gnomAD |
|
| rs1326741833 | 110 | H>P | No |
TOPMed gnomAD |
|
| rs1326741833 | 110 | H>R | No |
TOPMed gnomAD |
|
| rs75248193 | 110 | H>Y | No |
ExAC gnomAD |
|
| rs778063900 | 111 | T>A | No |
ExAC gnomAD |
|
| rs1232446318 | 111 | T>I | No | TOPMed | |
| rs2050881643 | 112 | E>K | No | gnomAD | |
| rs1325818727 | 113 | E>D | No |
TOPMed gnomAD |
|
| rs74751981 | 114 | K>N | No | Ensembl | |
| rs932129900 | 115 | Y>C | No |
TOPMed gnomAD |
|
| rs1273642700 | 123 | D>A | No |
TOPMed gnomAD |
|
| rs1273642700 | 123 | D>G | No |
TOPMed gnomAD |
|
| rs1432928058 | 124 | N>S | No | gnomAD | |
| rs1346327714 | 126 | M>I | No |
TOPMed gnomAD |
|
| rs1198270794 | 126 | M>R | No |
TOPMed gnomAD |
|
| rs1198270794 | 126 | M>T | No |
TOPMed gnomAD |
|
| rs75705863 | 127 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM1257576 rs376743625 |
127 | R>W | oesophagus [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1215628266 | 129 | H>Q | No |
TOPMed gnomAD |
|
| rs1176514813 | 129 | H>R | No | gnomAD | |
|
rs769315281 COSM3706467 |
130 | K>N | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs370659482 | 132 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1244309168 | 133 | P>S | No |
TOPMed gnomAD |
|
| rs1244309168 | 133 | P>T | No |
TOPMed gnomAD |
|
| rs755601081 | 134 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs755601081 | 134 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs748952164 | 137 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs768364064 | 138 | Q>H | No |
ExAC gnomAD |
|
| rs2153643619 | 139 | N>D | No | Ensembl | |
| rs774135500 | 140 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1470745947 | 140 | F>S | No |
TOPMed gnomAD |
|
| rs1479398678 | 142 | M>V | No |
TOPMed gnomAD |
|
| rs1427865641 | 143 | S>C | No |
TOPMed gnomAD |
|
| rs1165776215 | 145 | T>A | No |
TOPMed gnomAD |
|
| rs1165776215 | 145 | T>P | No |
TOPMed gnomAD |
|
| rs748024361 | 147 | P>T | No |
ExAC gnomAD |
|
| rs2051499353 | 148 | V>M | No | Ensembl | |
| rs1380907499 | 150 | S>I | No | gnomAD | |
| rs2051500614 | 151 | P>L | No | TOPMed | |
| rs1443951476 | 153 | A>T | No | gnomAD | |
| rs374058145 | 156 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs374058145 | 156 | Y>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs978256971 | 157 | T>A | No | TOPMed | |
| rs1255530789 | 157 | T>I | No |
TOPMed gnomAD |
|
| rs1198858305 | 159 | P>A | No | TOPMed | |
| rs2051505627 | 160 | G>A | No | Ensembl | |
| rs1391705773 | 161 | S>G | No |
TOPMed gnomAD |
|
| rs2051507511 | 161 | S>N | No | gnomAD | |
| rs765267108 | 162 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs2051508769 | 163 | L>V | No |
TOPMed gnomAD |
|
| rs764364084 | 164 | V>A | No |
ExAC gnomAD |
|
| rs554077925 | 164 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs554077925 | 164 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2051513864 | 166 | P>Q | No | TOPMed | |
| rs1327564911 | 166 | P>S | No |
TOPMed gnomAD |
|
| rs2051514573 | 167 | S>C | No | Ensembl | |
| rs1382046463 | 168 | L>W | No | gnomAD | |
| rs751746032 | 170 | A>D | No |
ExAC gnomAD |
|
| rs2051516643 | 172 | S>A | No | TOPMed | |
| rs189538320 | 173 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1360645449 | 173 | T>M | No |
TOPMed gnomAD |
|
| rs1271934645 | 177 | S>L | No | gnomAD | |
| rs1328517031 | 177 | S>T | No | gnomAD | |
| rs750924361 | 178 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs756576129 | 179 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs756576129 | 179 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs779722671 | 180 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1181704886 | 181 | S>F | No | gnomAD | |
| rs754532977 | 186 | T>P | No |
ExAC gnomAD |
|
| rs1406276041 | 187 | L>S | No | TOPMed | |
| rs772196260 | 187 | L>V | No |
TOPMed gnomAD |
|
| rs370083199 | 188 | H>L | No |
ESP gnomAD |
|
| rs1455405534 | 190 | N>H | No | TOPMed | |
| rs2051534278 | 190 | N>I | No | Ensembl | |
| rs2051535001 | 191 | V>M | No | Ensembl | |
| rs2051535972 | 192 | S>T | No | TOPMed | |
| rs890163554 | 195 | A>T | No |
TOPMed gnomAD |
|
| rs771790695 | 196 | P>T | No |
ExAC gnomAD |
|
| rs1339326152 | 197 | Q>K | No | gnomAD | |
| rs373867888 | 200 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1268801908 | 201 | S>N | No | gnomAD | |
| rs1364444037 | 202 | T>I | No |
TOPMed gnomAD |
|
| rs776542455 | 204 | N>S | No |
ExAC gnomAD |
|
| rs761954497 | 208 | M>T | No |
ExAC gnomAD |
|
| rs199820037 | 210 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs773300399 | 211 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs773300399 | 211 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs1376719387 | 212 | T>A | No | Ensembl | |
| rs1410768695 | 213 | D>E | No | gnomAD | |
| rs1567406543 | 213 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1420767777 | 214 | L>V | No | gnomAD | |
| rs2051778219 | 215 | T>A | No |
1000Genomes TOPMed gnomAD |
|
| rs2051778970 | 215 | T>I | No | Ensembl | |
| rs761164074 | 218 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs761164074 | 218 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs766830478 | 218 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs754420352 | 219 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs754420352 | 219 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs377207410 | 220 | A>S | No | Ensembl | |
| rs2051784633 | 220 | A>V | No | TOPMed | |
| rs1597067730 | 221 | G>A | No | Ensembl | |
| rs2051785378 | 221 | G>R | No | Ensembl | |
| rs752197525 | 223 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs757836374 | 224 | P>R | No |
ExAC gnomAD |
|
| rs2051788351 | 224 | P>T | No | Ensembl | |
|
COSM1300953 COSM1300952 COSM1300954 rs1338970568 |
226 | G>E | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs763844927 | 226 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1020821508 | 228 | G>E | No | Ensembl | |
| rs1314741371 | 229 | F>C | No | Ensembl | |
| rs1252757538 | 230 | V>G | No | gnomAD | |
| rs2055112442 | 230 | V>L | No | TOPMed | |
| rs2055113927 | 231 | N>H | No | gnomAD | |
| rs1458236440 | 231 | N>S | No | gnomAD | |
| rs2153715505 | 232 | S>P | No | Ensembl | |
| rs1021756347 | 233 | R>G | No |
TOPMed gnomAD |
|
| rs751251460 | 235 | S>Y | No |
ExAC gnomAD |
|
| rs966621137 | 237 | N>D | No | Ensembl | |
| rs966621137 | 237 | N>H | No | Ensembl | |
| rs1391618912 | 238 | L>F | No |
TOPMed gnomAD |
|
| rs756832092 | 239 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1368740492 | 239 | I>S | No |
TOPMed gnomAD |
|
| rs1368740492 | 239 | I>T | No |
TOPMed gnomAD |
|
| rs767312497 | 241 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2055122002 | 242 | T>A | No | gnomAD | |
|
COSM3401580 rs967480004 COSM3401581 COSM3401579 |
243 | G>D | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs750276293 | 245 | N>D | No |
ExAC TOPMed gnomAD |
|
| rs750276293 | 245 | N>H | No |
ExAC TOPMed gnomAD |
|
| rs755896449 | 246 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs780077974 | 246 | S>R | No |
ExAC gnomAD |
|
| rs189526729 | 248 | G>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1313179108 | 249 | K>E | No | gnomAD | |
| rs758530238 | 250 | V>G | No |
ExAC gnomAD |
|
| rs2055130350 | 251 | M>I | No | TOPMed | |
| rs966199515 | 251 | M>V | No |
TOPMed gnomAD |
|
| rs1318750476 | 252 | P>R | No | gnomAD | |
| rs199751824 | 253 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs199751824 | 253 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs919428393 | 254 | K>E | No |
TOPMed gnomAD |
|
| rs776983135 | 257 | P>A | No |
ExAC gnomAD |
|
| rs776983135 | 257 | P>S | No |
ExAC gnomAD |
|
| rs1201466862 | 258 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2055135601 | 259 | P>L | No | TOPMed | |
| rs2055136098 | 261 | G>D | No | Ensembl | |
| rs1261513727 | 262 | G>D | No | gnomAD | |
| rs746305206 | 262 | G>S | No |
ExAC gnomAD |
|
| rs1387648248 | 263 | N>K | No | gnomAD | |
| rs1190640339 | 264 | L>F | No | gnomAD | |
| rs2055140197 | 265 | G>E | No | TOPMed | |
| rs79454361 | 266 | M>L | No | Ensembl | |
| rs2055142592 | 268 | S>N | No | TOPMed | |
| rs2055141988 | 268 | S>R | No | Ensembl | |
| rs776085239 | 271 | P>R | No |
ExAC gnomAD |
|
| rs2055146525 | 272 | D>A | No | TOPMed | |
| rs1403197596 | 276 | V>I | No | gnomAD | |
| rs1467527861 | 278 | P>S | No |
TOPMed gnomAD |
|
|
COSM3747965 rs373219260 COSM3747966 COSM3747964 |
279 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs373219260 | 279 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373219260 | 279 | P>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2055153779 | 280 | S>L | No | TOPMed | |
| rs2055155186 | 281 | S>G | No | gnomAD | |
| rs2055155903 | 281 | S>N | No | Ensembl | |
| rs766041224 | 281 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs1281469976 | 282 | K>N | No | gnomAD | |
| rs754806380 | 285 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs2055159295 | 286 | P>A | No | Ensembl | |
| rs777926012 | 287 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs751751585 | 287 | P>R | No |
ExAC gnomAD |
|
| rs777926012 | 287 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs757410498 | 288 | L>P | No |
ExAC gnomAD |
|
| rs2153798349 | 289 | S>L | No | 1000Genomes | |
| rs1156456873 | 289 | S>P | No | TOPMed | |
| rs371903721 | 290 | E>K | No |
ESP TOPMed gnomAD |
|
| rs371903721 | 290 | E>Q | No |
ESP TOPMed gnomAD |
|
| rs1220030211 | 291 | E>K | No | gnomAD | |
| rs1278287218 | 293 | E>D | No | gnomAD | |
| rs1280625694 | 294 | L>F | No | gnomAD | |
| rs370231805 | 294 | L>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs2057650939 | 295 | E>G | No | TOPMed | |
| rs325408 | 297 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1597314532 | 298 | T>A | No | Ensembl | |
| rs1597314532 | 298 | T>P | No | Ensembl | |
| rs325407 | 299 | Q>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776198742 | 301 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1470079428 | 303 | S>I | No | gnomAD | |
| rs977516251 | 304 | S>Y | No | Ensembl | |
| rs745490285 | 305 | Q>K | No |
ExAC gnomAD |
|
| rs2058084124 | 306 | A>G | No | TOPMed | |
| rs926044570 | 307 | T>I | No | Ensembl | |
| rs769713418 | 307 | T>P | No |
ExAC gnomAD |
|
| rs986385216 | 309 | P>S | No |
TOPMed gnomAD |
|
| rs1403151791 | 310 | L>F | No |
TOPMed gnomAD |
|
| rs2058085784 | 310 | L>P | No |
TOPMed gnomAD |
|
| rs2153812291 | 312 | T>P | No | Ensembl | |
| rs774313312 | 313 | P>Q | No |
ExAC gnomAD |
|
| rs760715529 | 314 | V>A | No |
ExAC gnomAD |
|
|
COSM699660 COSM699661 rs766432411 COSM699659 |
315 | V>M | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1220727361 | 318 | T>I | No |
TOPMed gnomAD |
|
| rs754039795 | 319 | T>A | No |
ExAC gnomAD |
|
| rs755127907 | 319 | T>I | No |
ExAC gnomAD |
|
| rs755127907 | 319 | T>N | No |
ExAC gnomAD |
|
| rs2058088946 | 320 | P>A | No | TOPMed | |
| rs1432885682 | 322 | L>F | No | TOPMed | |
| rs753119776 | 323 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs778187283 | 324 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1177372159 | 328 | V>G | No | gnomAD | |
| rs1883840625 | 328 | V>M | No | Ensembl | |
| rs373204094 | 332 | M>L | No |
ESP TOPMed gnomAD |
|
| rs373204094 | 332 | M>V | No |
ESP TOPMed gnomAD |
|
| rs769625421 | 336 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs775274782 | 337 | N>D | No |
ExAC gnomAD |
|
| rs2153812563 | 337 | N>S | No | Ensembl | |
| rs752008306 | 340 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs752008306 | 340 | Y>F | No |
ExAC TOPMed gnomAD |
|
| rs1304465099 | 340 | Y>N | No | TOPMed | |
| rs752008306 | 340 | Y>S | No |
ExAC TOPMed gnomAD |
|
| rs751045780 | 343 | T>N | No | gnomAD | |
| rs751045780 | 343 | T>S | No | gnomAD | |
| rs373128826 | 345 | A>T | No |
TOPMed gnomAD |
|
| rs1167637574 | 345 | A>V | No |
TOPMed gnomAD |
|
| rs945679983 | 346 | D>V | No | Ensembl | |
| rs889147027 | 347 | L>P | No | TOPMed | |
| rs1171751474 | 348 | S>* | No |
TOPMed gnomAD |
|
| rs1425183121 | 349 | A>G | No |
TOPMed gnomAD |
|
| rs779671375 | 349 | A>S | No |
ExAC gnomAD |
|
|
COSM1375646 rs1425183121 COSM1375648 COSM1375647 |
349 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs754602729 | 352 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1293924463 | 353 | F>C | No | gnomAD | |
| rs778416281 | 354 | N>S | No |
ExAC TOPMed gnomAD |
|
|
rs748043464 COSM1375651 COSM1375649 COSM1375650 |
355 | S>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1359583377 | 356 | P>R | No | gnomAD | |
| rs777739995 | 357 | G>E | No |
ExAC gnomAD |
|
| rs746903028 | 360 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1314445937 | 362 | G>R | No | gnomAD | |
| rs2153825201 | 363 | Q>* | No | 1000Genomes | |
| rs775485813 | 363 | Q>R | No |
ExAC gnomAD |
|
| rs749311515 | 364 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs768987862 | 365 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs2058549110 | 366 | A>S | No | gnomAD | |
| rs774402211 | 366 | A>V | No |
ExAC gnomAD |
|
| rs762285015 | 368 | Q>K | No |
ExAC gnomAD |
|
| rs2058549972 | 370 | H>R | No | gnomAD | |
| rs767723022 | 371 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs773886574 | 373 | G>A | No |
ExAC gnomAD |
|
| rs761196788 | 375 | A>T | No |
ExAC gnomAD |
|
| rs1199433031 | 376 | A>V | No | gnomAD | |
| rs766002593 | 378 | S>N | No |
ExAC TOPMed gnomAD |
|
|
COSM86677 rs1037830845 |
379 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs897873312 | 379 | S>Y | No | TOPMed | |
| rs1160720970 | 381 | V>A | No | gnomAD | |
| rs1597353539 | 382 | A>G | No | Ensembl | |
| rs1378845081 | 382 | A>S | No | gnomAD | |
| rs886153299 | 383 | G>A | No |
TOPMed gnomAD |
|
| rs2058737971 | 384 | G>R | No |
TOPMed gnomAD |
|
| rs763506462 | 385 | Q>* | No | gnomAD | |
| rs763506462 | 385 | Q>K | No | gnomAD | |
| rs2058739142 | 387 | S>P | No | TOPMed | |
| rs2058739403 | 388 | Q>P | No | TOPMed | |
| rs2058739663 | 389 | G>C | No | TOPMed | |
| rs2058739945 | 390 | S>Y | No | Ensembl | |
| rs375108417 | 391 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1323038646 | 392 | L>V | No |
TOPMed gnomAD |
|
| rs1056672385 | 393 | S>C | No |
TOPMed gnomAD |
|
| rs1397523154 | 393 | S>T | No |
TOPMed gnomAD |
|
| rs1456397524 | 396 | T>S | No | gnomAD | |
| rs1200413525 | 397 | N>H | No | gnomAD | |
| rs776974654 | 397 | N>I | No |
ExAC gnomAD |
|
| rs2058741986 | 398 | Q>K | No | TOPMed | |
| rs1012602620 | 400 | I>V | No |
TOPMed gnomAD |
|
| rs2058743396 | 401 | S>I | No |
TOPMed gnomAD |
|
| rs2058743396 | 401 | S>N | No |
TOPMed gnomAD |
|
| rs2058743702 | 404 | S>F | No | TOPMed | |
| rs2058744428 | 405 | E>K | No | gnomAD | |
| rs2058744685 | 406 | P>L | No | gnomAD | |
| rs1476923080 | 410 | P>R | No |
TOPMed gnomAD |
|
|
rs752428663 COSM3936692 |
411 | R>Q | oesophagus [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs764606745 | 411 | R>W | No |
ExAC gnomAD |
|
| rs1490237826 | 412 | D>Y | No | TOPMed | |
| rs372533601 | 413 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372533601 | 413 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs763893561 | 414 | M>I | No | ExAC | |
|
COSM1608129 rs1332579803 |
414 | M>V | liver [Cosmic] | No |
cosmic curated gnomAD |
| rs1359000862 | 415 | T>I | No |
TOPMed gnomAD |
|
| rs1359000862 | 415 | T>S | No |
TOPMed gnomAD |
|
| rs2058748452 | 416 | P>L | No | TOPMed | |
| rs369579124 | 417 | S>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1007445184 | 418 | G>C | No | gnomAD | |
| rs1007445184 | 418 | G>R | No | gnomAD | |
| rs2058750888 | 419 | F>L | No | Ensembl | |
| rs866679051 | 419 | F>L | No | gnomAD | |
| rs1381368214 | 420 | Q>H | No | gnomAD | |
| rs2058754590 | 420 | Q>R | No | TOPMed | |
| rs2058756201 | 421 | Q>H | No | Ensembl | |
| rs2058756475 | 422 | Q>P | No | TOPMed | |
| rs1597354773 | 424 | Q>P | No | Ensembl | |
| rs1597354773 | 424 | Q>R | No | Ensembl | |
| rs200861006 | 425 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs755114794 | 426 | Q>* | No | ExAC | |
| rs757465493 | 426 | Q>P | No | Ensembl | |
| rs2058761037 | 428 | Q>E | No | TOPMed | |
| rs975766668 | 428 | Q>H | No |
TOPMed gnomAD |
|
| rs965675950 | 428 | Q>P | No | Ensembl | |
|
rs3138597 CA7755653 RCV000455204 |
429 | Q>missing | No |
ClinGen ClinVar dbSNP |
|
| rs201861701 | 429 | Q>P | No | gnomAD | |
| rs1193162148 | 430 | Q>H | No |
TOPMed gnomAD |
|
| rs199811207 | 430 | Q>P | No | Ensembl | |
| rs778791945 | 431 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs778791945 | 431 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs778791945 | 431 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs369815961 | 432 | P>L | No |
1000Genomes ESP TOPMed gnomAD |
|
|
rs369815961 COSM321649 |
432 | P>Q | lung [Cosmic] | No |
cosmic curated 1000Genomes ESP TOPMed gnomAD |
| rs369815961 | 432 | P>R | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs1157233240 | 433 | P>Q | No |
TOPMed gnomAD |
|
| rs1169041802 | 433 | P>S | No | gnomAD | |
| rs1052360469 | 434 | P>L | No |
TOPMed gnomAD |
|
| rs1052360469 | 434 | P>Q | No |
TOPMed gnomAD |
|
| rs1052360469 | 434 | P>R | No |
TOPMed gnomAD |
|
|
TCGA novel rs1487767106 |
435 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1487767106 | 435 | P>Q | No |
TOPMed gnomAD |
|
| rs1487767106 | 435 | P>R | No |
TOPMed gnomAD |
|
| rs778137086 | 435 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs778137086 | 435 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs2153832754 | 436 | Q>H | No | Ensembl | |
| rs2153832749 | 436 | Q>P | No | Ensembl | |
| rs2153832749 | 436 | Q>R | No | Ensembl | |
| rs1218337915 | 437 | P>A | No | Ensembl | |
| rs1218337915 | 437 | P>S | No | Ensembl | |
| rs2058779860 | 438 | Q>P | No | Ensembl | |
| rs1337825692 | 440 | Q>H | No | gnomAD | |
| rs2058780506 | 440 | Q>P | No | gnomAD | |
| VAR_017743 | 440 | Q>del | loss of nuclear localization; 66% decrease in transcription activation; loss of synergistic activation by MEF2A and GATA1 through a dominant-negative mechanism [UniProt] | No | UniProt |
| rs2058781849 | 441 | P>L | No | Ensembl | |
| rs1290730801 | 441 | P>S | No | TOPMed | |
| rs527482416 | 442 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1344231640 | 443 | Q>H | No | gnomAD | |
| rs2153832890 | 443 | Q>P | No | Ensembl | |
| rs2058784217 | 445 | Q>H | No | Ensembl | |
| rs1286041381 | 445 | Q>P | No | TOPMed | |
| rs1286041381 | 445 | Q>R | No | TOPMed | |
| rs2058784558 | 446 | P>S | No | TOPMed | |
| rs1280505902 | 447 | R>* | No | gnomAD | |
| rs769355862 | 447 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs769355862 | 447 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs769355862 | 447 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2058786070 | 448 | Q>K | No | TOPMed | |
| rs1284113826 | 449 | E>K | No | gnomAD | |
| rs2153833008 | 450 | M>K | No | Ensembl | |
| rs762442500 | 452 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs762442500 | 452 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1246144047 | 452 | R>H | No |
TOPMed gnomAD |
|
| rs1246144047 | 452 | R>L | No |
TOPMed gnomAD |
|
| rs1246144047 | 452 | R>P | No |
TOPMed gnomAD |
|
| rs1477329333 | 454 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1193389239 | 455 | V>G | No | gnomAD | |
| rs370921147 | 457 | S>G | No |
ESP ExAC gnomAD |
|
| rs1419448991 | 457 | S>I | No | gnomAD | |
| rs370921147 | 457 | S>R | No |
ESP ExAC gnomAD |
|
| rs751187554 | 459 | S>G | No | ExAC | |
| rs1361613790 | 459 | S>N | No |
TOPMed gnomAD |
|
| rs1157745214 | 461 | S>C | No | gnomAD | |
| rs761616362 | 463 | S>G | No | ExAC | |
| rs1358216124 | 463 | S>N | No | gnomAD | |
| rs1689357946 | 465 | Y>C | No | TOPMed | |
| rs1318408228 | 466 | D>G | No | gnomAD | |
| rs2058792780 | 467 | G>A | No | TOPMed | |
| rs1329960155 | 468 | S>G | No |
TOPMed gnomAD |
|
| rs1276526406 | 470 | R>Q | No |
TOPMed gnomAD |
|
| rs750377523 | 470 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1371515606 | 471 | E>K | No | gnomAD | |
| rs2058794449 | 473 | P>A | No | Ensembl | |
| rs780027711 | 474 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs755946369 | 474 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1356142229 | 475 | G>D | No | gnomAD | |
| rs1289284711 | 476 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs758521525 | 479 | S>F | No |
ExAC TOPMed |
|
| rs758521525 | 479 | S>Y | No |
ExAC TOPMed |
|
| rs1224480388 | 480 | P>L | No |
TOPMed gnomAD |
|
| rs1224480388 | 480 | P>Q | No |
TOPMed gnomAD |
|
| rs2153833300 | 480 | P>S | No | Ensembl | |
| rs771456542 | 481 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs2058798382 | 481 | I>T | No | Ensembl | |
|
rs111748677 RCV000960069 |
481 | I>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs997173174 | 482 | V>A | No |
TOPMed gnomAD |
|
| rs2058799232 | 482 | V>M | No | TOPMed | |
| rs1597357106 | 484 | G>S | No | Ensembl | |
|
COSM5182641 rs1470974611 |
485 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs370778860 | 485 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1317422523 | 486 | P>H | No | TOPMed | |
| rs868004044 | 487 | P>Q | No | Ensembl | |
| rs1403069029 | 487 | P>S | No | gnomAD | |
| rs1327924467 | 488 | N>D | No | gnomAD | |
| rs2058803328 | 491 | D>G | No | gnomAD | |
| rs571138739 | 493 | E>* | No |
1000Genomes ExAC gnomAD |
|
| rs2058803608 | 493 | E>D | No | gnomAD | |
| rs571138739 | 493 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs1445884204 | 494 | S>C | No |
TOPMed gnomAD |
|
| rs1445884204 | 494 | S>G | No |
TOPMed gnomAD |
|
| rs1306816502 | 497 | V>I | No | gnomAD | |
| rs2058805082 | 499 | R>* | No | Ensembl | |
| rs770266173 | 499 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs768102176 | 504 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs950518601 | 504 | A>V | No |
TOPMed gnomAD |
|
| rs761387435 | 505 | W>* | No |
ExAC gnomAD |
|
| rs767358305 | 505 | W>C | No |
ExAC gnomAD |
|
| rs1240434415 | 508 | T>E | No | gnomAD |
1 associated diseases with Q02078
[MIM: 608320]: Coronary artery disease, autosomal dominant, 1 (ADCAD1)
A common heart disease characterized by reduced or absent blood flow in one or more of the arteries that encircle and supply the heart. Its most important complication is acute myocardial infarction. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A common heart disease characterized by reduced or absent blood flow in one or more of the arteries that encircle and supply the heart. Its most important complication is acute myocardial infarction. Note=The disease is caused by variants affecting the gene represented in this entry.
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| transcription regulator complex | A protein complex that is capable of associating with DNA by direct binding, or via other DNA-binding proteins or complexes, and regulating transcription. |
14 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| histone acetyltransferase binding | Binding to an histone acetyltransferase. |
| histone deacetylase binding | Binding to histone deacetylase. |
| protein heterodimerization activity | Binding to a nonidentical protein to form a heterodimer. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | Binding to a specific sequence of DNA that is part of a regulatory region that controls the transcription of a gene or cistron by RNA polymerase II. |
| RNA polymerase II-specific DNA-binding transcription factor binding | Binding to a sequence-specific DNA binding RNA polymerase II transcription factor, any of the factors that interact selectively and non-covalently with a specific DNA sequence in order to modulate transcription. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| SMAD binding | Binding to a SMAD signaling protein. |
18 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cardiac conduction | Transfer of an organized electrical impulse across the heart to coordinate the contraction of cardiac muscles. The process begins with generation of an action potential (in the sinoatrial node (SA) in humans) and ends with a change in the rate, frequency, or extent of the contraction of the heart muscles. |
| cell differentiation | The cellular developmental process in which a relatively unspecialized cell, e.g. embryonic or regenerative cell, acquires specialized structural and/or functional features that characterize a specific cell. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cellular response to calcium ion | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a calcium ion stimulus. |
| dendrite morphogenesis | The process in which the anatomical structures of a dendrite are generated and organized. |
| DNA-templated transcription | The synthesis of an RNA transcript from a DNA template. |
| ERK5 cascade | An intracellular protein kinase cascade containing at least ERK5 (also called BMK1; a MAPK), a MEK (a MAPKK) and a MAP3K. The cascade can also contain an additional tier |
| heart development | The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers |
| mitochondrial genome maintenance | The maintenance of the structure and integrity of the mitochondrial genome; includes replication and segregation of the mitochondrial chromosome. |
| mitochondrion distribution | Any process that establishes the spatial arrangement of mitochondria between and within cells. |
| muscle organ development | The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of cardiac muscle hypertrophy | Any process that increases the rate, frequency or extent of the enlargement or overgrowth of all or part of the heart due to an increase in size (not length) of individual cardiac muscle fibers, without cell division. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of glucose import | Any process that activates or increases the frequency, rate or extent of the import of the hexose monosaccharide glucose into a cell or organelle. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| ventricular cardiac myofibril assembly | The process whose specific outcome is the progression of the ventricular cardiac myofibril over time, from its formation to the mature structure. A cardiac myofibril is a myofibril specific to cardiac muscle cells. |
17 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VDZ3 | MEF2A | Myocyte-specific enhancer factor 2A | Bos taurus (Bovine) | SS |
| Q9W6U8 | MEF2A | Myocyte-specific enhancer factor 2A | Gallus gallus (Chicken) | SS |
| Q06413 | MEF2C | Myocyte-specific enhancer factor 2C | Homo sapiens (Human) | EV |
| Q8CFN5 | Mef2c | Myocyte-specific enhancer factor 2C | Mus musculus (Mouse) | SS |
| Q60929 | Mef2a | Myocyte-specific enhancer factor 2A | Mus musculus (Mouse) | SS |
| A4UTP7 | MEF2C | Myocyte-specific enhancer factor 2C | Sus scrofa (Pig) | SS |
| A2ICN5 | MEF2A | Myocyte-specific enhancer factor 2A | Sus scrofa (Pig) | SS |
| A0A096MJY4 | Mef2c | Myocyte-specific enhancer factor 2C | Rattus norvegicus (Rat) | SS |
| Q2MJT0 | Mef2a | Myocyte-specific enhancer factor 2A | Rattus norvegicus (Rat) | SS |
| Q40702 | MADS2 | MADS-box transcription factor 2 | Oryza sativa subsp japonica (Rice) | PR |
| Q5K4R0 | MADS47 | MADS-box transcription factor 47 | Oryza sativa subsp japonica (Rice) | PR |
| Q655V4 | MADS30 | MADS-box transcription factor 30 | Oryza sativa subsp japonica (Rice) | PR |
| P35632 | AP3 | Floral homeotic protein APETALA 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P29383 | AGL3 | Agamous-like MADS-box protein AGL3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q683D7 | MAF5 | Protein MADS AFFECTING FLOWERING 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q1PFA4 | AGL30 | Agamous-like MADS-box protein AGL30 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FUY6 | J | MADS-box protein JOINTLESS | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGRKKIQITR | IMDERNRQVT | FTKRKFGLMK | KAYELSVLCD | CEIALIIFNS | SNKLFQYAST |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DMDKVLLKYT | EYNEPHESRT | NSDIVEALNK | KEHRGCDSPD | PDTSYVLTPH | TEEKYKKINE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EFDNMMRNHK | IAPGLPPQNF | SMSVTVPVTS | PNALSYTNPG | SSLVSPSLAA | SSTLTDSSML |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SPPQTTLHRN | VSPGAPQRPP | STGNAGGMLS | TTDLTVPNGA | GSSPVGNGFV | NSRASPNLIG |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ATGANSLGKV | MPTKSPPPPG | GGNLGMNSRK | PDLRVVIPPS | SKGMMPPLSE | EEELELNTQR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ISSSQATQPL | ATPVVSVTTP | SLPPQGLVYS | AMPTAYNTDY | SLTSADLSAL | QGFNSPGMLS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LGQVSAWQQH | HLGQAALSSL | VAGGQLSQGS | NLSINTNQNI | SIKSEPISPP | RDRMTPSGFQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| QQQQQQQQQQ | PPPPPQPQPQ | PPQPQPRQEM | GRSPVDSLSS | SSSSYDGSDR | EDPRGDFHSP |
| 490 | 500 | ||||
| IVLGRPPNTE | DRESPSVKRM | RMDAWVT |