Q01814
Gene name |
ATP2B2 (PMCA2) |
Protein name |
Plasma membrane calcium-transporting ATPase 2 |
Names |
PMCA2 , EC 7.2.2.10 , Plasma membrane calcium ATPase isoform 2 , Plasma membrane calcium pump isoform 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:491 |
EC number |
7.2.2.10: Linked to the hydrolysis of a nucleoside triphosphate |
Protein Class |
|
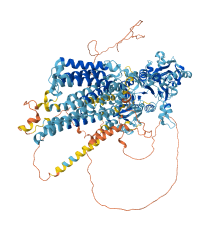
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
380-1086 (P-type ATPase domain) |
Relief mechanism |
Partner binding, Ligand binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q01814
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q01814-F1 | Predicted | AlphaFoldDB |
837 variants for Q01814
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs2124844270 RCV002051600 |
319 | A>missing | Hearing loss, autosomal dominant 82 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001250408 rs2061464108 |
345 | Q>* | Autosomal dominant nonsyndromic hearing loss [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV003335365 CA16617794 rs1064795322 RCV000484854 |
457 | E>K | ATP2B2-related disorder [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1487836380 RCV002539744 RCV001723265 |
503 | T>M | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
|
RCV000019379 CA127431 VAR_084465 rs61736451 RCV000954243 RCV001258253 |
631 | V>M | Agenesis of the corpus callosum with peripheral neuropathy Agenesis of the corpus callosum with peripheral neuropathy (accpn) Deafness, autosomal recessive 12, modifier of may exacerbate the severity of non-syndromic sensorineural hearing loss in patients with clinically relevant CDH23 variants; resulted in 50% decrease of the calcium ATPase activity [ClinVar, Ensembl, UniProt] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD UniProt |
|
rs267599520 RCV002051607 |
655 | E>* | Hearing loss, autosomal dominant 82 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
| VAR_086987 | 655 | E>del | DFNA82 [UniProt] | Yes | UniProt |
|
rs752990722 RCV002051608 |
666 | C>* | Hearing loss, autosomal dominant 82 [ClinVar] | Yes |
ClinVar ExAC TOPMed dbSNP gnomAD |
| VAR_086988 | 666 | C>del | DFNA82 [UniProt] | Yes | UniProt |
|
RCV002051601 rs2060537889 |
777 | R>* | Hearing loss, autosomal dominant 82 [ClinVar] | Yes |
ClinVar TOPMed dbSNP |
| VAR_086989 | 777 | R>del | DFNA82 [UniProt] | Yes | UniProt |
|
RCV000431659 CA16604786 rs1057520233 RCV002265755 |
922 | S>L | ATP2B2-related Progressive hearing impairment [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
| rs199652706 | 2 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs199652706 | 2 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs370454484 | 2 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1251132863 | 4 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
|
RCV001375144 rs1473080344 |
4 | M>V | No |
ClinVar dbSNP gnomAD |
|
| rs367841742 | 5 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs773766088 | 6 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs770275289 | 7 | S>N | No |
ExAC gnomAD |
|
| rs1257724369 | 7 | S>R | No | gnomAD | |
| rs564187066 | 8 | D>N | No |
1000Genomes ExAC gnomAD |
|
| rs777173772 | 10 | Y>C | No |
ExAC gnomAD |
|
| rs955039798 | 11 | S>P | No | Ensembl | |
| rs1311937271 | 12 | K>E | No | Ensembl | |
| rs768994822 | 13 | N>I | No |
ExAC gnomAD |
|
| rs768994822 | 13 | N>T | No |
ExAC gnomAD |
|
| rs2125180555 | 15 | R>G | No | Ensembl | |
| rs202179800 | 15 | R>T | No | 1000Genomes | |
|
RCV001925500 rs779940389 |
18 | S>L | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs779940389 | 18 | S>W | No |
ExAC TOPMed gnomAD |
|
| rs746087232 | 19 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs2063958294 | 19 | S>R | No | Ensembl | |
| rs778723997 | 20 | H>D | No |
ExAC gnomAD |
|
| rs375463057 | 20 | H>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs375463057 | 20 | H>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs777800527 | 22 | G>V | No |
ExAC gnomAD |
|
| rs1191658662 | 23 | E>A | No |
TOPMed gnomAD |
|
| rs759325739 | 23 | E>D | No |
ExAC gnomAD |
|
| rs752261081 | 23 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs765743831 | 25 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs762290443 | 27 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs776820559 | 27 | T>I | No |
ExAC gnomAD |
|
| rs776820559 | 27 | T>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs376205166 | 28 | M>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2063956078 | 28 | M>T | No |
TOPMed gnomAD |
|
| rs376205166 | 28 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1575260398 | 30 | E>A | No | Ensembl | |
| rs2063955969 | 30 | E>Q | No | gnomAD | |
|
COSM3584840 COSM3584841 rs761073766 |
32 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1012015300 | 32 | R>H | No | gnomAD | |
| rs1012015300 | 32 | R>L | No | gnomAD | |
| rs1236193426 | 34 | L>F | No | gnomAD | |
|
COSM3845719 rs775707408 |
38 | R>Q | breast [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
rs1423976084 COSM1593165 COSM1035932 |
38 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed |
| rs772109445 | 39 | G>A | No |
ExAC gnomAD |
|
| rs772109445 | 39 | G>D | No |
ExAC gnomAD |
|
| rs1381294391 | 39 | G>S | No | gnomAD | |
| rs746140832 | 40 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs2063954104 | 41 | E>K | No | Ensembl | |
| rs2125179844 | 43 | V>L | No | Ensembl | |
| rs771105825 | 44 | V>A | No |
ExAC gnomAD |
|
| rs771105825 | 44 | V>G | No |
ExAC gnomAD |
|
| rs779131837 | 44 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs1167932512 | 45 | K>E | No |
TOPMed gnomAD |
|
| rs1369127683 | 46 | I>M | No | gnomAD | |
| rs201490012 | 48 | E>G | No | gnomAD | |
| rs2063952940 | 48 | E>K | No | TOPMed | |
| rs777369218 | 49 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs556799597 | 50 | Y>* | No |
1000Genomes ExAC gnomAD |
|
|
rs756130380 RCV002010997 |
50 | Y>F | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs939364959 | 50 | Y>H | No | Ensembl | |
| rs902475656 | 51 | G>E | No | Ensembl | |
| rs1247238992 | 51 | G>W | No | gnomAD | |
| rs2063951607 | 52 | D>N | No | Ensembl | |
| rs893603911 | 53 | T>I | No | TOPMed | |
| rs1367491508 | 53 | T>P | No | TOPMed | |
| rs2063951110 | 54 | E>K | No | Ensembl | |
| rs751064368 | 56 | I>T | No |
ExAC gnomAD |
|
| rs766159464 | 57 | C>G | No |
ExAC gnomAD |
|
| rs1357836291 | 57 | C>S | No | gnomAD | |
| rs1357836291 | 57 | C>Y | No | gnomAD | |
| rs1280488968 | 58 | R>Q | No |
TOPMed gnomAD |
|
| rs762455685 | 58 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1364888558 | 59 | R>C | No |
TOPMed gnomAD |
|
|
rs149328739 COSM1536068 |
59 | R>H | lung central_nervous_system [Cosmic] | No |
cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
| rs149328739 | 59 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772521297 | 61 | K>I | No |
ExAC TOPMed gnomAD |
|
| rs1370993889 | 61 | K>Q | No | gnomAD | |
| rs772521297 | 61 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1271003547 | 63 | S>T | No |
TOPMed gnomAD |
|
| rs759648002 | 64 | P>R | No |
ExAC gnomAD |
|
| rs2063948925 | 65 | V>G | No | TOPMed | |
| rs1456485863 | 66 | E>G | No |
TOPMed gnomAD |
|
| rs201471815 | 68 | L>F | No |
1000Genomes ExAC gnomAD |
|
| rs768020427 | 69 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs768020427 | 69 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1377790802 | 69 | P>S | No | gnomAD | |
| rs1253332335 | 70 | G>D | No |
TOPMed gnomAD |
|
| rs1253332335 | 70 | G>V | No |
TOPMed gnomAD |
|
|
COSM6033401 COSM6033400 rs370301558 |
72 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1276267982 | 73 | P>S | No | Ensembl | |
| rs1202494397 | 74 | D>A | No | gnomAD | |
| rs1346124536 | 74 | D>E | No | gnomAD | |
| rs763274394 | 74 | D>Y | No |
ExAC gnomAD |
|
| rs2062584823 | 75 | L>P | No | Ensembl | |
| rs773281872 | 76 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1438701018 | 78 | R>S | No | gnomAD | |
| rs953578883 | 79 | K>R | No |
TOPMed gnomAD |
|
| rs747925050 | 82 | F>L | No |
ExAC gnomAD |
|
| rs776500526 | 84 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs746939018 | 86 | F>I | No |
ExAC TOPMed gnomAD |
|
| rs746939018 | 86 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs746939018 | 86 | F>V | No |
ExAC TOPMed gnomAD |
|
| rs779753195 | 87 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs757826245 | 88 | P>L | No |
ExAC gnomAD |
|
| rs2125018047 | 89 | P>Q | No | 1000Genomes | |
| rs745337447 | 94 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs745337447 | 94 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1413724494 | 96 | L>P | No | gnomAD | |
| rs753077209 | 99 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs138368540 | 99 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1559301935 | 101 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
|
COSM3673795 rs2062582481 COSM3673796 |
102 | A>V | Variant assessed as Somatic; MODERATE impact. prostate [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs766526597 | 106 | V>M | No |
ExAC gnomAD |
|
| rs762962570 | 107 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs41293357 | 112 | E>* | No | Ensembl | |
| rs776635799 | 113 | I>L | No |
ExAC gnomAD |
|
| rs1401643778 | 113 | I>T | No | TOPMed | |
| rs776635799 | 113 | I>V | No |
ExAC gnomAD |
|
| rs2062580990 | 114 | A>T | No | gnomAD | |
| rs760746487 | 115 | A>S | No |
ExAC TOPMed gnomAD |
|
|
COSM1417526 rs760746487 COSM1417527 |
115 | A>T | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1396454512 | 116 | I>V | No | Ensembl | |
| rs775591897 | 117 | I>V | No |
ExAC gnomAD |
|
| rs1011997168 | 122 | S>T | No | TOPMed | |
| rs779821154 | 123 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs745377655 | 123 | F>L | No |
ExAC gnomAD |
|
| rs2062578318 | 124 | Y>* | No | TOPMed | |
| rs2062579317 | 124 | Y>H | No | Ensembl | |
|
rs2125017372 RCV001961645 |
125 | H>Q | No |
ClinVar Ensembl dbSNP |
|
| rs1182921959 | 126 | P>L | No |
TOPMed gnomAD |
|
| rs1405158756 | 126 | P>S | No |
TOPMed gnomAD |
|
| rs781572230 | 128 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs375034217 | 129 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs755311598 | 129 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs780455268 | 130 | G>D | No |
ExAC gnomAD |
|
| rs1190277487 | 130 | G>S | No |
TOPMed gnomAD |
|
|
COSM3584830 COSM3584831 rs758872197 |
132 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs750481711 | 133 | G>R | No |
ExAC gnomAD |
|
| rs753931034 | 135 | A>T | No |
ExAC TOPMed gnomAD |
|
|
COSM1593166 rs945844574 COSM1035931 |
135 | A>V | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs756350386 | 136 | T>K | No |
ExAC TOPMed gnomAD |
|
|
COSM2916366 COSM2916367 rs756350386 |
136 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs767344205 | 137 | A>V | No |
ExAC gnomAD |
|
| rs759427038 | 139 | G>S | No |
ExAC gnomAD |
|
| rs774319358 | 140 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs774319358 | 140 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs151303929 | 141 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2062249436 | 143 | D>G | No | TOPMed | |
| rs1251848571 | 145 | G>R | No |
TOPMed gnomAD |
|
| rs1203494572 | 147 | A>V | No | gnomAD | |
| rs2124972512 | 149 | A>T | No | Ensembl | |
| rs769447734 | 152 | I>V | No |
ExAC gnomAD |
|
| rs1575127686 | 153 | E>G | No | Ensembl | |
|
COSM2916353 COSM2916354 rs1575127646 |
155 | A>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs775785242 | 155 | A>P | No |
ExAC gnomAD |
|
| rs775785242 | 155 | A>T | No |
ExAC gnomAD |
|
| rs1342769421 | 156 | A>T | No |
TOPMed gnomAD |
|
| rs2124972284 | 156 | A>V | No | Ensembl | |
| rs1298086139 | 159 | L>F | No | gnomAD | |
| rs2062247663 | 162 | I>S | No | Ensembl | |
|
rs2062247663 RCV001765388 |
162 | I>T | No |
ClinVar Ensembl dbSNP |
|
| rs757774695 | 163 | C>Y | No |
ExAC gnomAD |
|
|
COSM3786801 COSM3786802 rs749396633 |
168 | T>M | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1472971663 | 171 | N>S | No | gnomAD | |
| rs777990801 | 172 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1478665529 | 176 | E>D | No | TOPMed | |
| rs975345684 | 179 | F>L | No | TOPMed | |
|
COSM1417522 rs1479653862 COSM1417523 |
180 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
RCV002226138 COSM3720213 rs2124971839 |
180 | R>W | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ClinVar Ensembl dbSNP |
| rs1322308785 | 184 | S>N | No | gnomAD | |
|
COSM6051854 COSM6051855 rs754821593 |
185 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1417521 COSM1417520 rs766306375 |
187 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs772766333 | 193 | T>I | No |
ExAC gnomAD |
|
| rs776369165 | 194 | V>L | No |
ExAC TOPMed gnomAD |
|
|
COSM4616875 rs776369165 COSM4616876 |
194 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs367698989 | 196 | R>Q | No |
ESP ExAC gnomAD |
|
| rs2062244427 | 196 | R>W | No |
TOPMed gnomAD |
|
| rs1575127227 | 197 | A>S | No | Ensembl | |
| rs1458001350 | 199 | Q>E | No | gnomAD | |
| rs774802099 | 201 | V>D | No |
ExAC gnomAD |
|
| rs984601396 | 201 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs749812718 | 202 | Q>H | No |
ExAC gnomAD |
|
|
RCV002269473 rs2124971330 |
204 | P>A | No |
ClinVar Ensembl dbSNP |
|
| rs2062243234 | 204 | P>L | No | TOPMed | |
| rs2062242893 | 206 | A>D | No | Ensembl | |
| rs189987955 | 206 | A>P | No |
1000Genomes TOPMed gnomAD |
|
| rs189987955 | 206 | A>S | No |
1000Genomes TOPMed gnomAD |
|
| rs1183922750 | 207 | E>A | No | gnomAD | |
| rs1460690008 | 208 | I>F | No | gnomAD | |
| rs1486024532 | 209 | V>A | No | gnomAD | |
| rs145312453 | 209 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1323990065 | 210 | V>A | No | gnomAD | |
| rs781505875 | 210 | V>I | No |
ExAC gnomAD |
|
| rs1245948160 | 211 | G>R | No | gnomAD | |
| rs751381940 | 213 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs374835840 | 214 | A>S | No |
ESP ExAC gnomAD |
|
| rs374835840 | 214 | A>T | No |
ESP ExAC gnomAD |
|
| rs2124970907 | 218 | Y>N | No | Ensembl | |
| rs1575124532 | 220 | D>A | No | Ensembl | |
| rs77470961 | 221 | L>P | No | Ensembl | |
| rs141093862 | 223 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
|
rs758211761 TCGA novel |
225 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC gnomAD |
|
COSM5491627 rs2062203038 COSM5491626 |
225 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1434611666 | 226 | G>S | No | gnomAD | |
|
rs1475754485 RCV001945839 |
234 | L>V | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs758879461 | 235 | K>R | No | Ensembl | |
| rs535964962 | 236 | I>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs535964962 | 236 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778828310 | 236 | I>V | No |
ExAC gnomAD |
|
| rs753448558 | 237 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1356717742 | 240 | S>P | No |
TOPMed gnomAD |
|
| rs1352544972 | 248 | V>A | No | TOPMed | |
|
COSM1417518 COSM1417519 rs1057261655 |
249 | R>C | ovary Variant assessed as Somatic; MODERATE impact. large_intestine [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1057261655 | 249 | R>G | No | gnomAD | |
| rs200591536 | 249 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs375973812 COSM4111992 COSM258761 |
252 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP NCI-TCGA TOPMed gnomAD |
| rs1185431696 | 253 | D>N | No | TOPMed | |
| rs2062199903 | 254 | K>N | No | TOPMed | |
| rs1241347106 | 256 | P>A | No |
TOPMed gnomAD |
|
| rs372187689 | 263 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
RCV001905877 rs2124875506 |
264 | V>L | No |
ClinVar Ensembl dbSNP |
|
|
COSM1471478 rs775610841 |
270 | R>W | prostate [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1321099025 | 271 | M>I | No | Ensembl | |
| rs767551489 | 271 | M>L | No |
ExAC gnomAD |
|
| rs534464297 | 274 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs1405062514 | 275 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2061745238 | 278 | V>A | No | Ensembl | |
| rs771149479 | 278 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2061744926 | 284 | I>T | No | Ensembl | |
| rs1010759595 | 284 | I>V | No |
TOPMed gnomAD |
|
| rs769912544 | 285 | I>V | No |
ExAC gnomAD |
|
| rs768259928 | 290 | G>R | No |
ExAC gnomAD |
|
| rs779577877 | 291 | A>S | No |
ExAC gnomAD |
|
| rs1481610801 | 291 | A>V | No | gnomAD | |
| rs1435165880 | 292 | G>S | No |
TOPMed gnomAD |
|
|
rs367699943 VAR_084464 RCV002114326 |
293 | G>S | may exacerbate the severity of non-syndromic sensorineural hearing loss in patients with clinically relevant CDH23 variants; delayed calcium export [UniProt] | No |
ClinVar ESP ExAC TOPMed dbSNP gnomAD UniProt |
| rs754211616 | 294 | E>K | No |
ExAC gnomAD |
|
|
RCV000443086 CA16604434 rs373192330 |
295 | E>D | No |
TOPMed gnomAD ClinGen ClinVar dbSNP |
|
| rs778104787 | 295 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1170059047 | 300 | D>E | No | gnomAD | |
|
COSM3914442 COSM3914441 rs1355294301 |
300 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs753177348 | 301 | K>R | No |
ExAC gnomAD |
|
| rs956959764 | 303 | G>D | No |
TOPMed gnomAD |
|
| rs956959764 | 303 | G>V | No |
TOPMed gnomAD |
|
| rs2061684431 | 304 | V>L | No | TOPMed | |
| rs925504101 | 306 | K>R | No | Ensembl | |
| rs2061684217 | 307 | G>R | No | Ensembl | |
| rs768532843 | 308 | D>G | No |
ExAC gnomAD |
|
| rs2061684142 | 308 | D>N | No | Ensembl | |
| rs2061684008 | 310 | L>F | No | TOPMed | |
| rs1280329991 | 310 | L>R | No |
TOPMed gnomAD |
|
| rs760656114 | 312 | L>P | No |
ExAC gnomAD |
|
| rs1297964063 | 313 | P>R | No | gnomAD | |
| rs1383029083 | 314 | A>T | No |
TOPMed gnomAD |
|
| rs770633275 | 315 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs748937189 | 316 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1197959106 | 316 | D>G | No |
TOPMed gnomAD |
|
| rs147881294 | 317 | G>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs147881294 | 317 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs147881294 | 317 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs769045116 | 317 | G>V | No |
ExAC gnomAD |
|
|
rs747289941 COSM5732754 |
318 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1225625317 | 320 | A>G | No | gnomAD | |
| rs758766390 | 320 | A>T | No |
ExAC gnomAD |
|
| rs989534248 | 322 | N>I | No |
TOPMed gnomAD |
|
| rs1668281837 | 323 | A>D | No | TOPMed | |
| rs1282797967 | 325 | D>E | No | gnomAD | |
| rs1324369555 | 325 | D>G | No | gnomAD | |
| rs2061642296 | 326 | S>C | No | TOPMed | |
|
rs149708084 COSM1163318 |
327 | A>V | liver Variant assessed as Somatic; MODERATE impact. pancreas endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs974822079 | 328 | N>S | No | Ensembl | |
| rs757592586 | 329 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs554208140 | 330 | S>R | No | Ensembl | |
| rs754089404 | 331 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs1391779442 | 331 | L>V | No | gnomAD | |
| rs2061641423 | 334 | G>C | No | gnomAD | |
| rs1182334568 | 336 | M>V | No | TOPMed | |
| rs2061465089 | 337 | Q>P | No | TOPMed | |
| rs890878049 | 339 | G>D | No | Ensembl | |
| rs751479685 | 340 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs2061464563 | 341 | V>M | No | TOPMed | |
| rs762483506 | 342 | D>N | No |
ExAC gnomAD |
|
| rs181008668 | 343 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1451248706 | 344 | S>I | No |
TOPMed gnomAD |
|
| rs1575066214 | 344 | S>R | No | Ensembl | |
| rs1402690266 | 348 | A>T | No |
TOPMed gnomAD |
|
| rs536636171 | 349 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs751122 | 350 | Q>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1264227687 | 352 | D>Y | No | gnomAD | |
| rs2061436557 | 353 | G>E | No | TOPMed | |
| rs1439146629 | 353 | G>R | No |
TOPMed gnomAD |
|
| rs1258281902 | 354 | A>G | No | gnomAD | |
| rs1258281902 | 354 | A>V | No | gnomAD | |
| rs2061436420 | 355 | A>P | No | Ensembl | |
| rs1240770392 | 356 | A>T | No |
TOPMed gnomAD |
|
| rs2061436201 | 357 | M>V | No | TOPMed | |
| rs758399805 | 364 | S>N | No |
ExAC gnomAD |
|
| rs745769945 | 364 | S>R | No |
ExAC gnomAD |
|
| rs1224594944 | 365 | A>G | No | gnomAD | |
| rs753319875 | 368 | G>D | No |
ExAC gnomAD |
|
| rs1444840288 | 368 | G>S | No |
TOPMed gnomAD |
|
| rs752033239 | 369 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1464317425 | 369 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs766822562 | 370 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs766822562 | 370 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1559253285 | 370 | A>V | No | gnomAD | |
| rs1424406166 | 372 | D>N | No | gnomAD | |
| rs1189455392 | 375 | K>N | No | gnomAD | |
| rs1471023838 | 376 | A>D | No | gnomAD | |
| rs145244597 | 376 | A>S | No | TOPMed | |
| rs145244597 | 376 | A>T | No | TOPMed | |
| rs1471023838 | 376 | A>V | No | gnomAD | |
| rs765738283 | 377 | S>N | No |
ExAC gnomAD |
|
| rs761803682 | 380 | K>R | No |
ExAC gnomAD |
|
| rs776634300 | 383 | K>M | No |
ExAC gnomAD |
|
| rs150203000 | 385 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs150203000 | 385 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs991634566 | 390 | L>R | No | Ensembl | |
| rs1326253064 | 398 | G>W | No | TOPMed | |
| rs1374495991 | 400 | A>E | No | gnomAD | |
| rs1410901563 | 407 | I>L | No | gnomAD | |
| rs2061359799 | 408 | T>M | No | TOPMed | |
| rs2061359613 | 409 | V>A | No | TOPMed | |
| rs1488876496 | 410 | I>V | No | gnomAD | |
| rs146495987 | 415 | Y>S | No | ESP | |
| rs891845763 | 417 | T>S | No |
TOPMed gnomAD |
|
| rs2061359206 | 417 | T>S | No | Ensembl | |
| rs2061359080 | 418 | V>M | No |
TOPMed gnomAD |
|
| rs1244257141 | 420 | T>N | No |
TOPMed gnomAD |
|
| rs757802757 | 420 | T>P | No |
ExAC TOPMed gnomAD |
|
| rs757802757 | 420 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1297759850 | 422 | V>G | No |
TOPMed gnomAD |
|
| rs368504026 | 422 | V>M | No |
ExAC gnomAD |
|
| rs1030374767 | 424 | N>K | No | TOPMed | |
| rs2061358635 | 425 | K>Q | No | Ensembl | |
| rs149899981 | 427 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs149899981 | 427 | P>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs764543689 | 427 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1575055192 | 432 | C>G | No | Ensembl | |
| rs774052464 | 432 | C>Y | No |
ExAC gnomAD |
|
| rs916921377 | 433 | T>A | No |
TOPMed gnomAD |
|
|
COSM1035923 rs770734675 COSM1154059 |
433 | T>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs374649471 | 435 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs747675057 | 437 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs768261203 | 439 | Y>F | No |
ExAC gnomAD |
|
| rs1209648649 | 440 | F>L | No | gnomAD | |
| rs2061357326 | 444 | F>S | No | gnomAD | |
|
rs1553564078 CA658657275 RCV000522327 |
444 | F>missing | No |
ClinGen ClinVar dbSNP |
|
| rs757647596 | 446 | I>T | No |
ExAC gnomAD |
|
| rs758218985 | 446 | I>V | No |
ExAC TOPMed gnomAD |
|
|
rs1276448598 COSM1669967 COSM1669966 |
448 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
RCV001772583 rs2125521057 |
450 | V>L | No |
ClinVar Ensembl dbSNP |
|
| rs2061356468 | 455 | V>M | No | Ensembl | |
| rs766670443 | 463 | V>A | No |
ExAC gnomAD |
|
|
rs751832350 COSM3780614 |
463 | V>I | pancreas [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs1575054828 | 464 | T>P | No | Ensembl | |
| rs2061355722 | 465 | I>V | No | TOPMed | |
| rs1198836153 | 466 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
COSM3584813 rs1256194224 COSM3584812 |
470 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs771774308 | 476 | K>N | No |
ExAC gnomAD |
|
| rs143304473 | 476 | K>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1299396343 | 477 | D>N | No | gnomAD | |
| rs1459287444 | 478 | N>H | No | TOPMed | |
|
rs1378929747 COSM3408086 |
482 | R>C | central_nervous_system [Cosmic] | No |
cosmic curated gnomAD |
|
COSM3391941 rs1272655222 |
482 | R>H | pancreas [Cosmic] | No |
cosmic curated gnomAD |
| rs1367903479 | 486 | A>T | No | gnomAD | |
| rs2061257569 | 490 | M>V | No | Ensembl | |
| rs2061257431 | 492 | N>D | No |
TOPMed gnomAD |
|
| rs1359976671 | 494 | T>I | No |
TOPMed gnomAD |
|
| rs748424364 | 496 | I>V | No |
ExAC gnomAD |
|
|
COSM6162837 rs1265214151 COSM6162838 COSM1536078 |
497 | C>S | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs768990611 | 499 | D>E | No |
ExAC gnomAD |
|
| rs780277141 | 508 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs758734872 | 508 | R>H | No |
ExAC gnomAD |
|
| rs2061256198 | 509 | M>V | No |
TOPMed gnomAD |
|
| rs771678072 | 512 | V>A | No |
ExAC TOPMed gnomAD |
|
| rs1283038342 | 512 | V>I | No |
TOPMed gnomAD |
|
| rs368441051 | 515 | Y>C | No |
ESP ExAC gnomAD |
|
| rs368441051 | 515 | Y>F | No |
ESP ExAC gnomAD |
|
| rs764150216 | 517 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1575044678 | 518 | D>A | No | Ensembl | |
|
RCV002213069 rs752239299 |
518 | D>N | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
COSM3780612 RCV002149240 rs144118750 |
519 | V>I | pancreas [Cosmic] | No |
cosmic curated ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
| rs758994103 | 520 | H>L | No |
ExAC TOPMed gnomAD |
|
| rs2061255030 | 520 | H>N | No | Ensembl | |
| rs758994103 | 520 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1000988843 | 521 | Y>C | No |
TOPMed gnomAD |
|
| rs1000988843 | 521 | Y>S | No |
TOPMed gnomAD |
|
| rs1477462213 | 522 | K>Q | No | gnomAD | |
|
COSM3584809 rs2061254577 COSM3584808 |
523 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs2125505389 | 524 | I>F | No | Ensembl | |
| rs904108983 | 524 | I>M | No |
TOPMed gnomAD |
|
| rs2061254423 | 525 | P>T | No | TOPMed | |
| rs1559242841 | 526 | D>E | No | Ensembl | |
| rs202118324 | 526 | D>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs202118324 | 526 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs769272435 | 528 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs769272435 | 528 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs2061253908 | 529 | S>A | No | gnomAD | |
| rs780332176 | 530 | I>V | No |
ExAC gnomAD |
|
| rs1218495158 | 531 | N>D | No | gnomAD | |
| rs972126032 | 532 | T>S | No |
TOPMed gnomAD |
|
| rs2061253512 | 534 | T>S | No | Ensembl | |
| rs2061253267 | 535 | M>T | No | TOPMed | |
|
rs745874947 RCV002033455 |
535 | M>V | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
| rs1559242748 | 537 | L>M | No | Ensembl | |
| rs1559242748 | 537 | L>V | No | Ensembl | |
| rs56120857 | 539 | I>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs56120857 | 539 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs199678871 | 540 | N>D | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3660061 rs1467431326 |
540 | N>S | liver [Cosmic] | No |
cosmic curated gnomAD |
| rs1401047704 | 542 | I>V | No | gnomAD | |
| rs777396418 | 543 | A>S | No |
ExAC TOPMed gnomAD |
|
|
rs777396418 RCV000422716 COSM4664342 COSM4664341 CA2253626 |
543 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs756122323 | 544 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs2061252222 | 546 | S>T | No | gnomAD | |
| rs1256110308 | 547 | A>T | No |
TOPMed gnomAD |
|
| rs921352439 | 548 | Y>C | No |
TOPMed gnomAD |
|
| rs2061251666 | 549 | T>A | No | TOPMed | |
| rs2061251588 | 549 | T>I | No | Ensembl | |
| rs973900247 | 553 | L>P | No |
TOPMed gnomAD |
|
| rs765779585 | 554 | P>S | No |
ExAC gnomAD |
|
| rs765779585 | 554 | P>T | No |
ExAC gnomAD |
|
| rs2060854574 | 558 | E>K | No | TOPMed | |
| rs750067208 | 559 | G>S | No |
ExAC gnomAD |
|
| rs761133287 | 560 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2060854133 | 562 | P>H | No | TOPMed | |
| rs768204746 | 563 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs768204746 | 563 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs760088594 | 564 | Q>E | No |
ExAC gnomAD |
|
| rs774535349 | 565 | V>G | No |
ExAC gnomAD |
|
| rs749344787 | 567 | N>S | No |
ExAC TOPMed gnomAD |
|
|
rs1314290874 COSM4749854 COSM4749855 |
569 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs554270950 | 570 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs199935477 RCV001932806 |
571 | C>* | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs780965218 | 572 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1219521271 | 576 | F>I | No | gnomAD | |
| rs200139854 | 577 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2060852323 | 583 | D>G | No | TOPMed | |
| rs1230695512 | 583 | D>Y | No | TOPMed | |
| rs1191448274 | 585 | E>D | No | Ensembl | |
| rs764853768 | 585 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs778129111 | 586 | P>H | No | gnomAD | |
| rs778129111 | 586 | P>L | No | gnomAD | |
| rs2125451022 | 586 | P>S | No | Ensembl | |
| rs759927011 | 587 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs759927011 RCV002005626 |
587 | V>M | No |
ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
COSM4054935 rs1360871132 COSM4054936 |
588 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs774748492 | 588 | R>H | No |
ExAC gnomAD |
|
| rs766839476 | 589 | S>C | No |
ExAC gnomAD |
|
| rs1559557507 | 590 | Q>L | No | Ensembl | |
| rs1165102345 | 592 | P>A | No | gnomAD | |
| rs2060850956 | 594 | E>K | No | TOPMed | |
| rs2060850792 | 597 | Y>F | No | TOPMed | |
| rs2060850716 | 599 | V>M | No | TOPMed | |
|
COSM1417512 COSM1417511 rs199991046 |
605 | V>M | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1593176 rs776463160 COSM1035918 |
606 | R>C | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs768463609 COSM3584804 COSM3584805 |
606 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
RCV001997084 rs2125450656 |
609 | M>T | No |
ClinVar Ensembl dbSNP |
|
| rs1440719706 | 610 | S>N | No |
TOPMed gnomAD |
|
| rs1440719706 | 610 | S>T | No |
TOPMed gnomAD |
|
| rs1220319437 | 611 | T>S | No | gnomAD | |
| rs2125450573 | 613 | I>M | No | Ensembl | |
| rs148749880 | 617 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
RCV000885465 rs150683478 |
617 | D>N | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1293738196 | 618 | E>D | No |
TOPMed gnomAD |
|
| rs778533707 | 618 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1274732075 | 619 | S>N | No |
TOPMed gnomAD |
|
|
rs757018981 COSM580772 COSM1143082 |
621 | R>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1391563891 | 621 | R>H | No |
TOPMed gnomAD |
|
| rs1391563891 | 621 | R>L | No |
TOPMed gnomAD |
|
| rs939205374 | 624 | S>N | No | Ensembl | |
| rs753563395 | 630 | I>V | No | ExAC | |
| rs61736451 | 631 | V>L | Agenesis of the corpus callosum with peripheral neuropathy (accpn) [Ensembl] | No |
1000Genomes ESP ExAC TOPMed gnomAD |
| rs2060848090 | 632 | L>F | No | Ensembl | |
| rs182847338 | 634 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs765698071 | 637 | K>Q | No |
ExAC gnomAD |
|
| rs1398265960 | 639 | L>F | No | gnomAD | |
| rs140327013 | 640 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1433214170 | 641 | G>R | No | gnomAD | |
|
COSM6162845 COSM6162846 rs753859888 COSM1536086 |
642 | A>E | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs753859888 | 642 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2060816186 | 643 | G>A | No | Ensembl | |
| rs2060816259 | 643 | G>R | No | TOPMed | |
| rs764364185 | 644 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs74592691 | 646 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA gnomAD |
| rs199797486 | 646 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373876706 | 648 | F>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs759212037 | 649 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs774434270 | 649 | R>P | No |
ExAC TOPMed gnomAD |
|
|
COSM1484389 rs774434270 COSM1484390 |
649 | R>Q | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs759212037 | 649 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1287918907 | 650 | P>L | No |
TOPMed gnomAD |
|
| rs1484557674 | 650 | P>S | No | gnomAD | |
| rs748864193 | 651 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs145574094 | 651 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs748864193 | 651 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs369920268 | 652 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1593177 rs758833343 |
653 | R>Q | endometrium [Cosmic] | No |
cosmic curated ExAC gnomAD |
|
COSM727477 COSM1149534 rs544333948 |
653 | R>W | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs746169153 | 654 | D>Y | No |
ExAC gnomAD |
|
| rs267599520 | 655 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2060814546 | 656 | M>I | No | gnomAD | |
| rs1476355125 | 658 | K>E | No |
TOPMed gnomAD |
|
| rs753956510 | 660 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs200255705 | 661 | I>M | No |
1000Genomes ExAC gnomAD |
|
| rs2060814219 | 661 | I>T | No | Ensembl | |
| rs1014421005 | 662 | E>K | No | Ensembl | |
| rs756094148 | 663 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs376966138 | 667 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs1281662606 | 672 | I>V | No |
TOPMed gnomAD |
|
| rs772900118 | 674 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1461301540 | 677 | R>H | No |
TOPMed gnomAD |
|
| rs1043392470 | 678 | D>N | No |
TOPMed gnomAD |
|
| rs1342958763 | 680 | P>S | No |
TOPMed gnomAD |
|
| rs1342958763 | 680 | P>T | No |
TOPMed gnomAD |
|
| rs1430417763 | 681 | S>G | No | gnomAD | |
| rs572312326 | 683 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2060812271 | 683 | P>S | No | Ensembl | |
|
RCV000997988 rs1575005369 |
684 | E>missing | No |
ClinVar dbSNP |
|
|
COSM3695736 rs746292256 COSM3695735 |
685 | P>L | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs746292256 | 685 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs757806989 | 686 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1406406151 | 689 | N>S | No | Ensembl | |
| rs749814396 | 690 | E>Q | No |
ExAC gnomAD |
|
| rs1215845724 | 691 | N>Y | No | gnomAD | |
| rs909716038 | 694 | L>F | No |
TOPMed gnomAD |
|
| rs553746671 | 695 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs752661102 COSM3584799 COSM3584798 |
696 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs767837099 | 699 | C>Y | No |
ExAC gnomAD |
|
|
COSM2916272 COSM2916271 rs1211251049 |
702 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
RCV001287954 rs2060810402 |
703 | V>L | No |
ClinVar Ensembl dbSNP |
|
| rs2060810244 | 704 | G>D | No | Ensembl | |
|
rs2060810244 RCV002248252 |
704 | G>V | No |
ClinVar Ensembl dbSNP |
|
|
RCV001765851 rs2125445636 |
705 | I>F | No |
ClinVar Ensembl dbSNP |
|
| rs1486066765 | 706 | E>K | No | TOPMed | |
|
rs751442343 COSM366419 |
708 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs760871469 | 711 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs567994556 | 712 | E>Q | No |
1000Genomes ExAC gnomAD |
|
| rs1468202061 | 715 | E>G | No |
TOPMed gnomAD |
|
| rs938764659 | 718 | R>C | No |
TOPMed gnomAD |
|
| rs928659205 | 718 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs200250553 | 722 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs773443414 | 722 | R>W | No |
ExAC gnomAD |
|
| rs1188002884 | 724 | G>D | No | gnomAD | |
| rs1317767271 | 724 | G>S | No | gnomAD | |
|
rs1574980625 RCV000991464 |
725 | I>N | No |
ClinVar Ensembl dbSNP |
|
| rs748582045 | 726 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs748582045 | 726 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs754680596 | 728 | R>C | No |
TOPMed gnomAD |
|
| rs780440699 | 728 | R>H | No |
ExAC gnomAD |
|
| rs780440699 | 728 | R>L | No |
ExAC gnomAD |
|
| rs1415335547 | 729 | M>V | No | gnomAD | |
|
rs750301862 COSM3766831 |
733 | D>N | liver [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs2060549734 | 735 | I>S | No | TOPMed | |
| rs1421091853 | 737 | T>K | No | gnomAD | |
| rs1421091853 | 737 | T>M | No | gnomAD | |
| rs778544762 | 739 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs778544762 | 739 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1345399209 | 739 | R>W | No | gnomAD | |
| rs1398451336 | 740 | A>D | No | gnomAD | |
| rs757024697 | 741 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1354662233 | 742 | A>S | No |
TOPMed gnomAD |
|
|
COSM1184062 rs1354662233 |
742 | A>T | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2060548868 | 747 | I>T | No | Ensembl | |
| rs755630251 | 749 | H>L | No |
ExAC TOPMed gnomAD |
|
| rs755630251 | 749 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1559545069 | 749 | H>Y | No | Ensembl | |
| rs1441382565 | 754 | F>I | No |
TOPMed gnomAD |
|
| rs1323514737 | 756 | C>F | No |
TOPMed gnomAD |
|
| rs759078548 | 758 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs773673140 | 760 | K>R | No |
ExAC gnomAD |
|
| rs1291420073 | 765 | R>K | No | gnomAD | |
| rs762199117 | 767 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs749393972 | 767 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs749393972 | 767 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs762199117 | 767 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs747147456 | 769 | E>A | No |
ExAC gnomAD |
|
| rs747147456 | 769 | E>G | No |
ExAC gnomAD |
|
| rs964598436 | 769 | E>K | No |
TOPMed gnomAD |
|
| rs1469025797 | 770 | K>N | No | Ensembl | |
| rs1195731524 | 771 | G>R | No |
TOPMed gnomAD |
|
| rs764534165 | 773 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs761076118 | 777 | R>L | No |
ExAC gnomAD |
|
|
rs761076118 COSM3584794 COSM3584795 |
777 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1382206628 | 779 | D>E | No |
TOPMed gnomAD |
|
| rs768007228 | 784 | K>Q | No |
ExAC gnomAD |
|
| rs1158776536 | 786 | R>Q | No |
TOPMed gnomAD |
|
| rs1574978966 | 787 | V>G | No | Ensembl | |
| rs2060537313 | 787 | V>L | No | TOPMed | |
| rs2060536914 | 790 | R>C | No |
TOPMed gnomAD |
|
| rs1280783124 | 790 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1209651936 | 791 | S>A | No | gnomAD | |
|
rs200946394 RCV001960397 |
794 | T>M | No |
ClinVar 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
| rs1334731513 | 795 | D>E | No |
TOPMed gnomAD |
|
| rs1574978828 | 796 | K>Q | No | Ensembl | |
| rs2125409089 | 802 | G>C | No | Ensembl | |
| rs781596200 | 805 | D>N | No |
ExAC gnomAD |
|
| rs969053907 | 807 | T>K | No | Ensembl | |
| rs1271169857 | 808 | H>Q | No |
TOPMed gnomAD |
|
| rs1341826559 | 808 | H>Y | No | gnomAD | |
| rs2060424412 | 809 | T>N | No | TOPMed | |
| rs2060424412 | 809 | T>S | No | TOPMed | |
| rs1179500847 | 810 | E>K | No | gnomAD | |
| rs758486263 | 812 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM2152966 COSM2152967 rs866132370 |
812 | R>W | Variant assessed as Somatic; MODERATE impact. central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs201856129 | 814 | V>G | No |
ExAC gnomAD |
|
| rs765512175 | 815 | V>G | No |
ExAC gnomAD |
|
|
rs1421342790 COSM3584788 COSM3584789 |
817 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
|
COSM6095865 COSM6095866 rs2060423534 COSM1143080 |
818 | T>M | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic Ensembl |
|
rs2125394967 RCV002273378 |
821 | G>R | No |
ClinVar Ensembl dbSNP |
|
| rs1251839245 | 825 | G>R | No | gnomAD | |
| rs1219954399 | 826 | P>T | No | gnomAD | |
| rs1440079928 | 827 | A>V | No | gnomAD | |
| rs2125394862 | 833 | V>L | No | Ensembl | |
| rs773964391 | 834 | G>S | No |
ExAC gnomAD |
|
| rs748989820 | 836 | A>T | No |
ExAC gnomAD |
|
| rs2060422192 | 837 | M>V | No | TOPMed | |
|
rs1485572860 RCV000762361 |
839 | I>N | No |
ClinVar dbSNP gnomAD |
|
| rs1485572860 | 839 | I>T | No | gnomAD | |
| rs1235942676 | 840 | A>T | No |
TOPMed gnomAD |
|
| rs746398574 | 853 | L>V | No |
ExAC gnomAD |
|
| rs2060405265 | 857 | N>D | No | Ensembl | |
| rs1354642256 | 860 | S>C | No | gnomAD | |
| rs111706887 | 861 | I>M | No |
TOPMed gnomAD |
|
|
COSM180598 rs778120026 COSM4111977 |
862 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1559538483 | 869 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs752585168 | 869 | R>L | No |
ExAC gnomAD |
|
| rs2060404279 | 871 | V>I | No | Ensembl | |
| rs1475079434 | 885 | V>I | No | gnomAD | |
| rs2060403610 | 886 | N>I | No | gnomAD | |
|
rs145040618 RCV001767817 |
886 | N>K | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM5863142 rs762583223 COSM5863141 RCV001977186 |
887 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs1206864634 | 891 | I>T | No | gnomAD | |
| rs2060403036 | 896 | G>V | No | gnomAD | |
|
rs566780843 COSM1245727 |
897 | A>T | oesophagus [Cosmic] | No |
cosmic curated 1000Genomes ExAC gnomAD |
| rs1027007294 | 898 | C>F | No |
TOPMed gnomAD |
|
| rs369741116 | 899 | I>V | No |
ESP ExAC TOPMed gnomAD |
|
|
rs1310353379 COSM1318746 COSM1318747 |
900 | T>M | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs2125380296 RCV002004121 |
904 | P>R | No |
ClinVar Ensembl dbSNP |
|
| rs2060322836 | 904 | P>T | No | Ensembl | |
| rs1273658754 | 908 | V>M | No | gnomAD | |
| rs2060322267 | 910 | M>I | No | TOPMed | |
| rs1574958820 | 913 | V>G | No | Ensembl | |
| rs1559534646 | 924 | A>T | No | Ensembl | |
| rs2060320930 | 927 | T>S | No | TOPMed | |
| rs760294184 | 929 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1328909942 | 929 | P>T | No | gnomAD | |
|
COSM1035914 rs368058047 COSM1154056 |
931 | T>M | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1384784896 | 932 | E>D | No | gnomAD | |
| rs954014031 | 933 | T>N | No | Ensembl | |
| rs2060319588 | 938 | K>E | No | Ensembl | |
|
RCV002265179 rs1559534482 |
939 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar NCI-TCGA dbSNP gnomAD |
| rs2060319515 | 939 | P>S | No | TOPMed | |
| rs2125379588 | 941 | G>D | No | Ensembl | |
| rs1419635133 | 942 | R>C | No | gnomAD | |
| rs780286157 | 942 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2060318884 | 943 | N>D | No | Ensembl | |
| rs1179707919 | 945 | P>L | No |
TOPMed gnomAD |
|
| rs1199537776 | 951 | M>I | No | gnomAD | |
| rs753392215 | 957 | G>S | No |
ExAC gnomAD |
|
| rs2060318011 | 960 | V>I | No | Ensembl | |
| rs755578943 | 966 | I>M | No |
ExAC gnomAD |
|
| rs1333691245 | 968 | T>I | No | gnomAD | |
| rs752295184 | 970 | L>F | No |
ExAC gnomAD |
|
| rs2060317186 | 972 | V>I | No | Ensembl | |
| rs1363070884 | 974 | E>K | No | gnomAD | |
| rs1015069977 | 975 | K>R | No | TOPMed | |
| rs2060249868 | 976 | M>I | No | TOPMed | |
| rs769765695 | 980 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs769765695 | 980 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs556727847 | 981 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1403379381 | 982 | G>E | No | gnomAD | |
| rs780731851 | 982 | G>R | No |
ExAC gnomAD |
|
|
RCV002011513 rs146069247 |
985 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ClinVar ESP ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs751257556 | 985 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1194938859 | 986 | P>A | No | gnomAD | |
|
rs865848016 COSM1578633 |
989 | S>L | meninges [Cosmic] | No |
cosmic curated Ensembl |
| rs2060248624 | 1001 | T>I | No |
TOPMed gnomAD |
|
|
COSM4111973 rs765963921 COSM4111974 |
1003 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs2060248337 | 1004 | M>V | No | TOPMed | |
| rs2060248037 | 1010 | E>K | No | TOPMed | |
| rs2060247971 | 1011 | I>V | No | gnomAD | |
| rs2060247845 | 1012 | N>S | No | TOPMed | |
| rs762801598 | 1013 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs772959601 | 1016 | I>M | No |
ExAC gnomAD |
|
|
RCV001878332 rs2125369003 |
1016 | I>V | No |
ClinVar Ensembl dbSNP |
|
| rs935376816 | 1017 | H>Y | No |
TOPMed gnomAD |
|
|
rs373150646 COSM4111972 COSM4111971 |
1018 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs2125368879 RCV001865033 |
1020 | R>missing | No |
ClinVar dbSNP |
|
| rs768217763 | 1020 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs760285222 | 1020 | R>H | No |
ExAC gnomAD |
|
|
RCV001945730 rs760285222 |
1020 | R>L | No |
ClinVar ExAC dbSNP gnomAD |
|
| rs779913030 | 1024 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2060246578 | 1025 | G>D | No | Ensembl | |
| rs758122334 | 1025 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs376018398 | 1026 | I>V | No |
ESP ExAC gnomAD |
|
| rs778300525 | 1028 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs753250088 | 1031 | I>N | No |
ExAC gnomAD |
|
| rs753250088 | 1031 | I>T | No |
ExAC gnomAD |
|
| rs2060246066 | 1033 | C>R | No | Ensembl | |
| rs1472307150 | 1036 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2060245415 | 1042 | I>T | No | Ensembl | |
| rs1229661551 | 1044 | I>T | No | TOPMed | |
|
rs1320953291 RCV001995070 |
1053 | P>S | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1287233328 | 1056 | C>S | No | gnomAD | |
| rs753542319 | 1057 | S>A | No |
ExAC gnomAD |
|
| rs764156405 | 1061 | L>P | No |
ExAC gnomAD |
|
| rs2060237724 | 1062 | D>N | No | TOPMed | |
| rs2060237575 | 1065 | M>L | No | Ensembl | |
| rs1432325485 | 1067 | C>G | No | gnomAD | |
| rs1432325485 | 1067 | C>S | No | gnomAD | |
| rs760673006 | 1068 | I>L | No |
ExAC gnomAD |
|
| rs1294701886 | 1068 | I>T | No |
TOPMed gnomAD |
|
| rs1348501104 | 1070 | I>T | No |
TOPMed gnomAD |
|
| rs1457666923 | 1070 | I>V | No | gnomAD | |
| rs2060236718 | 1075 | L>P | No | gnomAD | |
| rs2060236511 | 1076 | V>A | No | Ensembl | |
| rs149035567 | 1076 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2125367001 | 1077 | W>* | No | 1000Genomes | |
| rs754564501 | 1081 | I>M | No |
TOPMed gnomAD |
|
| rs149910726 | 1082 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1377740348 | 1083 | T>A | No | gnomAD | |
| rs370705388 | 1083 | T>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2060183530 | 1084 | I>T | No | Ensembl | |
| rs1559527736 | 1088 | R>T | No | Ensembl | |
| rs2060182748 | 1098 | L>F | No |
TOPMed gnomAD |
|
| rs1267513201 | 1099 | T>I | No | TOPMed | |
| rs868700600 | 1103 | E>K | No | Ensembl | |
| rs376285041 | 1104 | I>M | No |
ESP TOPMed gnomAD |
|
| rs2060182338 | 1104 | I>V | No | TOPMed | |
| rs778096122 | 1105 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs778096122 | 1105 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs1034835475 | 1106 | E>K | No | gnomAD | |
| rs752511520 | 1108 | E>K | No |
ExAC gnomAD |
|
| rs374011120 | 1110 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2060181384 | 1111 | E>D | No | TOPMed | |
| rs772535893 | 1111 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2060181106 | 1113 | V>A | No | TOPMed | |
| rs765812045 | 1113 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs765812045 | 1113 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1393017235 | 1114 | E>A | No | TOPMed | |
| rs1286872809 | 1117 | D>E | No | gnomAD | |
| rs1247509549 | 1118 | H>L | No | gnomAD | |
|
RCV001757917 rs2125357954 |
1119 | A>T | No |
ClinVar Ensembl dbSNP |
|
| rs1312365878 | 1119 | A>T | No |
TOPMed gnomAD |
|
| rs917336797 | 1119 | A>V | No |
TOPMed gnomAD |
|
|
RCV001752214 rs2125357897 |
1121 | R>P | No |
ClinVar Ensembl dbSNP |
|
| rs1267725418 | 1124 | R>Q | No |
TOPMed gnomAD |
|
|
COSM3391939 rs370895383 COSM3391940 |
1125 | R>Q | Variant assessed as Somatic; MODERATE impact. pancreas urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1457719867 | 1125 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2060179620 | 1126 | G>S | No | TOPMed | |
| rs1156558456 | 1131 | F>L | No |
TOPMed gnomAD |
|
|
rs2125357775 RCV001824499 |
1132 | R>* | No |
ClinVar Ensembl dbSNP |
|
| rs2060179332 | 1132 | R>Q | No | TOPMed | |
| rs1559527380 | 1135 | N>S | No | Ensembl | |
|
rs1421474714 RCV001771271 |
1136 | R>Q | No |
ClinVar dbSNP gnomAD |
|
| rs774850425 | 1139 | T>A | No |
ExAC gnomAD |
|
| rs1213345149 | 1142 | R>C | No | gnomAD | |
| rs779884532 | 1142 | R>H | No |
ExAC TOPMed gnomAD |
|
|
rs2059913127 COSM1133672 |
1143 | V>I | urinary_tract [Cosmic] | No |
cosmic curated TOPMed |
| rs373130153 | 1144 | V>M | No | ESP | |
| rs1328157604 | 1146 | A>V | No |
TOPMed gnomAD |
|
| rs1363944044 | 1148 | R>H | No | gnomAD | |
| rs1323903845 | 1152 | Y>C | No | TOPMed | |
| rs2059912422 | 1154 | G>D | No | Ensembl | |
| rs2059912489 | 1154 | G>R | No | TOPMed | |
| rs28533981 | 1158 | P>L | No | Ensembl | |
| rs972609276 | 1159 | E>D | No | Ensembl | |
| rs1430424392 | 1161 | R>L | No | gnomAD | |
| rs1430424392 | 1161 | R>Q | No | gnomAD | |
| rs1337909479 | 1162 | T>I | No |
TOPMed gnomAD |
|
| rs778779257 | 1163 | S>F | No |
ExAC gnomAD |
|
| rs2059911651 | 1168 | M>I | No | gnomAD | |
| rs1426632836 | 1168 | M>V | No | gnomAD | |
| rs1291305538 | 1169 | A>D | No |
TOPMed gnomAD |
|
| rs753318972 | 1170 | H>R | No |
ExAC gnomAD |
|
| rs1220151933 | 1170 | H>Y | No | gnomAD | |
| rs369789634 | 1172 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1417502 rs1215036750 |
1174 | R>Q | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1453823607 | 1174 | R>W | No |
TOPMed gnomAD |
|
| rs766773779 | 1175 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1206866742 | 1176 | E>K | No | gnomAD | |
| rs763370632 | 1177 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2059910816 | 1182 | I>N | No | gnomAD | |
| rs1167103851 | 1183 | P>S | No | TOPMed | |
| rs765817186 | 1185 | I>T | No |
ExAC gnomAD |
|
| rs2059910521 | 1186 | D>E | No | gnomAD | |
|
rs776766476 COSM1692367 |
1189 | D>N | skin [Cosmic] | No |
cosmic curated ExAC gnomAD |
| rs776766476 | 1189 | D>Y | No |
ExAC gnomAD |
|
| rs1344044122 | 1193 | D>H | No |
TOPMed gnomAD |
|
| rs1290397168 | 1193 | D>V | No | gnomAD | |
| rs2059910166 | 1194 | A>T | No | gnomAD | |
| rs768817061 | 1194 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs141356421 RCV002099811 |
1195 | A>T | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs577047176 | 1195 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1574915127 | 1199 | N>T | No | Ensembl | |
| rs1426967358 | 1200 | S>L | No |
TOPMed gnomAD |
|
| rs1426967358 | 1200 | S>W | No |
TOPMed gnomAD |
|
| rs2125319666 | 1201 | S>R | No | Ensembl | |
| rs1574915081 | 1201 | S>R | No | Ensembl | |
| rs1574915069 | 1201 | S>T | No | Ensembl | |
| rs1472694896 | 1202 | P>A | No | gnomAD | |
| rs953627979 | 1202 | P>L | No |
TOPMed gnomAD |
|
| rs2059908844 | 1204 | S>L | No | gnomAD | |
| rs1257045285 | 1207 | N>D | No | gnomAD | |
| rs2059908484 | 1212 | A>T | No | Ensembl | |
| rs780659874 | 1212 | A>V | No |
ExAC TOPMed gnomAD |
|
|
rs1432340467 COSM1154054 |
1214 | D>N | large_intestine endometrium [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2059908240 | 1216 | G>E | No | Ensembl | |
| rs1574914934 | 1218 | N>T | No | TOPMed | |
| rs1237796914 | 1219 | L>M | No | TOPMed | |
|
COSM1184060 rs765910999 |
1220 | T>M | large_intestine breast [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1574914845 | 1221 | T>P | No | Ensembl | |
| rs1472634369 | 1222 | D>E | No | gnomAD | |
| rs764236179 | 1222 | D>N | No |
ExAC gnomAD |
|
| rs1397790233 | 1224 | S>N | No |
TOPMed gnomAD |
|
| rs2059907467 | 1227 | A>G | No | TOPMed | |
| rs1337254392 | 1233 | G>R | No | gnomAD | |
| rs2059907033 | 1235 | P>A | No | Ensembl | |
| rs2059906964 | 1236 | I>F | No | gnomAD | |
| rs1454168225 | 1239 | L>P | No | gnomAD | |
| rs2059906743 | 1241 | T>M | No | Ensembl | |
| rs1293257670 | 1242 | S>* | No |
TOPMed gnomAD |
|
| rs1293257670 | 1242 | S>L | No |
TOPMed gnomAD |
2 associated diseases with Q01814
[MIM: 619804]: Deafness, autosomal dominant, 82 (DFNA82)
A form of non-syndromic, sensorineural hearing loss. Sensorineural hearing loss results from damage to the neural receptors of the inner ear, the nerve pathways to the brain, or the area of the brain that receives sound information. DNFA82 is characterized by onset of rapidly progressive bilateral sensorineural hearing loss usually early in the first decade. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of non-syndromic, sensorineural hearing loss. Sensorineural hearing loss results from damage to the neural receptors of the inner ear, the nerve pathways to the brain, or the area of the brain that receives sound information. DNFA82 is characterized by onset of rapidly progressive bilateral sensorineural hearing loss usually early in the first decade. . Note=The disease is caused by variants affecting the gene represented in this entry.
5 regional properties for Q01814
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cation-transporting P-type ATPase, N-terminal | 47 - 123 | IPR004014 |
| domain | Cation-transporting P-type ATPase, C-terminal | 903 - 1081 | IPR006068 |
| ptm | P-type ATPase, phosphorylation site | 499 - 505 | IPR018303 |
| domain | Plasma membrane calcium transporting P-type ATPase, C-terminal | 1126 - 1172 | IPR022141 |
| domain | P-type ATPase, haloacid dehalogenase domain | 479 - 870 | IPR044492 |
Functions
| Description | ||
|---|---|---|
| EC Number | 7.2.2.10 | Linked to the hydrolysis of a nucleoside triphosphate |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
13 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| basolateral plasma membrane | The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| dendritic spine membrane | The portion of the plasma membrane surrounding a dendritic spine. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| GABA-ergic synapse | A synapse that uses GABA as a neurotransmitter. These synapses are typically inhibitory. |
| glutamatergic synapse | A synapse that uses glutamate as a neurotransmitter. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| neuronal cell body membrane | The plasma membrane of a neuron cell body - excludes the plasma membrane of cell projections such as axons and dendrites. |
| parallel fiber to Purkinje cell synapse | An excitatory synapse formed by the parallel fibers of granule cells synapsing onto the dendrites of Purkinje cells. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density membrane | The membrane component of the postsynaptic density. This is the region of the postsynaptic membrane in which the population of neurotransmitter receptors involved in synaptic transmission are concentrated. |
| presynaptic active zone membrane | The membrane portion of the presynaptic active zone; it is the site where docking and fusion of synaptic vesicles occurs for the release of neurotransmitters. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction |
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| calmodulin binding | Binding to calmodulin, a calcium-binding protein with many roles, both in the calcium-bound and calcium-free states. |
| glutamate receptor binding | Binding to a glutamate receptor. |
| metal ion binding | Binding to a metal ion. |
| P-type calcium transporter activity | Enables the transfer of a solute or solutes from one side of a membrane to the other according to the reaction |
| P-type calcium transporter activity involved in regulation of postsynaptic cytosolic calcium ion concentration | A calcium-transporting P-type ATPase activity involved in regulation of postsynaptic cytosolic calcium ion concentration. |
| PDZ domain binding | Binding to a PDZ domain of a protein, a domain found in diverse signaling proteins. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| calcium ion transport | The directed movement of calcium (Ca) ions into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| monoatomic ion transmembrane transport | A process in which a monoatomic ion is transported across a membrane. Monatomic ions (also called simple ions) are ions consisting of exactly one atom. |
| neural retina development | The progression of the neural retina over time from its initial formation to the mature structure. The neural retina is the part of the retina that contains neurons and photoreceptor cells. |
| neuron differentiation | The process in which a relatively unspecialized cell acquires specialized features of a neuron. |
| regulation of cardiac conduction | Any process that modulates the frequency, rate or extent of cardiac conduction. |
| regulation of cytosolic calcium ion concentration | Any process involved in the maintenance of an internal steady state of calcium ions within the cytosol of a cell or between the cytosol and its surroundings. |
| sensory perception of sound | The series of events required for an organism to receive an auditory stimulus, convert it to a molecular signal, and recognize and characterize the signal. Sonic stimuli are detected in the form of vibrations and are processed to form a sound. |
31 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P38929 | PMC1 | Calcium-transporting ATPase 2 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| D3K0R6 | ATP2B4 | Plasma membrane calcium-transporting ATPase 4 | Bos taurus (Bovine) | SS |
| P23634 | ATP2B4 | Plasma membrane calcium-transporting ATPase 4 | Homo sapiens (Human) | EV |
| P20020 | ATP2B1 | Plasma membrane calcium-transporting ATPase 1 | Homo sapiens (Human) | SS |
| Q16720 | ATP2B3 | Plasma membrane calcium-transporting ATPase 3 | Homo sapiens (Human) | EV |
| G5E829 | Atp2b1 | Plasma membrane calcium-transporting ATPase 1 | Mus musculus (Mouse) | SS |
| Q6Q477 | Atp2b4 | Plasma membrane calcium-transporting ATPase 4 | Mus musculus (Mouse) | SS |
| Q9R0K7 | Atp2b2 | Plasma membrane calcium-transporting ATPase 2 | Mus musculus (Mouse) | SS |
| P23220 | ATP2B1 | Plasma membrane calcium-transporting ATPase 1 | Sus scrofa (Pig) | SS |
| Q64542 | Atp2b4 | Plasma membrane calcium-transporting ATPase 4 | Rattus norvegicus (Rat) | SS |
| P11505 | Atp2b1 | Plasma membrane calcium-transporting ATPase 1 | Rattus norvegicus (Rat) | SS |
| Q64568 | Atp2b3 | Plasma membrane calcium-transporting ATPase 3 | Rattus norvegicus (Rat) | SS |
| P11506 | Atp2b2 | Plasma membrane calcium-transporting ATPase 2 | Rattus norvegicus (Rat) | SS |
| Q8RUN1 | ACA1 | Calcium-transporting ATPase 1, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q2QY12 | ACA9 | Probable calcium-transporting ATPase 9, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q6ATV4 | ACA3 | Calcium-transporting ATPase 3, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q2QMX9 | ACA10 | Calcium-transporting ATPase 10, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q7XEK4 | ACA7 | Calcium-transporting ATPase 7, plasma membrane-type | Oryza sativa subsp japonica (Rice) | EV |
| Q7X8B5 | ACA5 | Calcium-transporting ATPase 5, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q2RAS0 | ACA8 | Probable calcium-transporting ATPase 8, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| Q65X71 | ACA6 | Probable calcium-transporting ATPase 6, plasma membrane-type | Oryza sativa subsp japonica (Rice) | SS |
| O22218 | ACA4 | Calcium-transporting ATPase 4, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M2L4 | ACA11 | Putative calcium-transporting ATPase 11, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O64806 | ACA7 | Putative calcium-transporting ATPase 7, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81108 | ACA2 | Calcium-transporting ATPase 2, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | EV |
| Q9LY77 | ACA12 | Calcium-transporting ATPase 12, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LIK7 | ACA13 | Putative calcium-transporting ATPase 13, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LU41 | ACA9 | Calcium-transporting ATPase 9, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LF79 | ACA8 | Calcium-transporting ATPase 8, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | EV |
| Q9SZR1 | ACA10 | Calcium-transporting ATPase 10, plasma membrane-type | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q37145 | ACA1 | Calcium-transporting ATPase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGDMTNSDFY | SKNQRNESSH | GGEFGCTMEE | LRSLMELRGT | EAVVKIKETY | GDTEAICRRL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KTSPVEGLPG | TAPDLEKRKQ | IFGQNFIPPK | KPKTFLQLVW | EALQDVTLII | LEIAAIISLG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LSFYHPPGEG | NEGCATAQGG | AEDEGEAEAG | WIEGAAILLS | VICVVLVTAF | NDWSKEKQFR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GLQSRIEQEQ | KFTVVRAGQV | VQIPVAEIVV | GDIAQVKYGD | LLPADGLFIQ | GNDLKIDESS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTGESDQVRK | SVDKDPMLLS | GTHVMEGSGR | MLVTAVGVNS | QTGIIFTLLG | AGGEEEEKKD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KKGVKKGDGL | QLPAADGAAA | SNAADSANAS | LVNGKMQDGN | VDASQSKAKQ | QDGAAAMEMQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PLKSAEGGDA | DDRKKASMHK | KEKSVLQGKL | TKLAVQIGKA | GLVMSAITVI | ILVLYFTVDT |
| 430 | 440 | 450 | 460 | 470 | 480 |
| FVVNKKPWLP | ECTPVYVQYF | VKFFIIGVTV | LVVAVPEGLP | LAVTISLAYS | VKKMMKDNNL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VRHLDACETM | GNATAICSDK | TGTLTTNRMT | VVQAYVGDVH | YKEIPDPSSI | NTKTMELLIN |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AIAINSAYTT | KILPPEKEGA | LPRQVGNKTE | CGLLGFVLDL | KQDYEPVRSQ | MPEEKLYKVY |
| 610 | 620 | 630 | 640 | 650 | 660 |
| TFNSVRKSMS | TVIKLPDESF | RMYSKGASEI | VLKKCCKILN | GAGEPRVFRP | RDRDEMVKKV |
| 670 | 680 | 690 | 700 | 710 | 720 |
| IEPMACDGLR | TICVAYRDFP | SSPEPDWDNE | NDILNELTCI | CVVGIEDPVR | PEVPEAIRKC |
| 730 | 740 | 750 | 760 | 770 | 780 |
| QRAGITVRMV | TGDNINTARA | IAIKCGIIHP | GEDFLCLEGK | EFNRRIRNEK | GEIEQERIDK |
| 790 | 800 | 810 | 820 | 830 | 840 |
| IWPKLRVLAR | SSPTDKHTLV | KGIIDSTHTE | QRQVVAVTGD | GTNDGPALKK | ADVGFAMGIA |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GTDVAKEASD | IILTDDNFSS | IVKAVMWGRN | VYDSISKFLQ | FQLTVNVVAV | IVAFTGACIT |
| 910 | 920 | 930 | 940 | 950 | 960 |
| QDSPLKAVQM | LWVNLIMDTF | ASLALATEPP | TETLLLRKPY | GRNKPLISRT | MMKNILGHAV |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| YQLALIFTLL | FVGEKMFQID | SGRNAPLHSP | PSEHYTIIFN | TFVMMQLFNE | INARKIHGER |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| NVFDGIFRNP | IFCTIVLGTF | AIQIVIVQFG | GKPFSCSPLQ | LDQWMWCIFI | GLGELVWGQV |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| IATIPTSRLK | FLKEAGRLTQ | KEEIPEEELN | EDVEEIDHAE | RELRRGQILW | FRGLNRIQTQ |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| IRVVKAFRSS | LYEGLEKPES | RTSIHNFMAH | PEFRIEDSQP | HIPLIDDTDL | EEDAALKQNS |
| 1210 | 1220 | 1230 | 1240 | ||
| SPPSSLNKNN | SAIDSGINLT | TDTSKSATSS | SPGSPIHSLE | TSL |