Q01534
Gene name |
TSPY1 (TSPY) |
Protein name |
Testis-specific Y-encoded protein 1 |
Names |
Cancer/testis antigen 78, CT78 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:728137 |
EC number |
|
Protein Class |
|
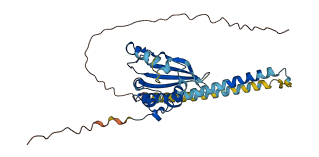
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for Q01534
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-Q01534-F1 | Predicted | AlphaFoldDB |
83 variants for Q01534
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA414973731 rs1603142470 |
4 | E>D | No |
ClinGen Ensembl |
|
|
CA414973735 rs1556182135 |
5 | G>D | No |
ClinGen Ensembl |
|
|
CA414974070 rs1603142511 |
50 | A>S | No |
ClinGen Ensembl |
|
| VAR_016226 | 79 | E>EVEVVAE | No | UniProt | |
|
rs1603142522 CA414974518 |
82 | G>S | No |
ClinGen Ensembl |
|
|
CA10572458 rs777840135 |
86 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA414974608 rs1482895379 |
89 | E>* | No |
ClinGen gnomAD |
|
|
CA10572459 rs765624506 |
90 | A>T | No |
ClinGen ExAC |
|
|
CA10572460 rs753013820 |
90 | A>V | No |
ClinGen ExAC |
|
|
CA10572461 rs758060583 |
92 | R>P | No |
ClinGen ExAC |
|
|
CA414974706 rs1233759717 |
97 | V>L | No |
ClinGen Ensembl |
|
|
CA414974723 rs1265245901 |
98 | P>R | No |
ClinGen gnomAD |
|
|
rs1185795405 CA414974718 |
98 | P>S | No |
ClinGen gnomAD |
|
|
rs1478212511 CA414974754 |
101 | G>R | No |
ClinGen gnomAD |
|
|
rs1193013751 CA414974786 |
103 | M>K | No |
ClinGen gnomAD |
|
|
rs1603142563 CA414974800 |
104 | T>N | No |
ClinGen Ensembl |
|
|
CA414974826 rs1421789884 |
106 | E>G | No |
ClinGen gnomAD |
|
|
CA414974827 rs1421789884 |
106 | E>V | No |
ClinGen gnomAD |
|
|
rs1416340243 CA414974840 |
107 | S>F | No |
ClinGen gnomAD |
|
|
CA414974843 rs1416340243 |
107 | S>Y | No |
ClinGen gnomAD |
|
|
CA414974849 rs1177249826 |
108 | A>S | No |
ClinGen gnomAD |
|
|
rs1358889000 CA414974867 |
109 | P>L | No |
ClinGen gnomAD |
|
|
CA10572462 rs777419549 |
112 | L>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1400128427 CA414974915 |
113 | L>Q | No |
ClinGen gnomAD |
|
|
CA414974930 rs1276963055 |
114 | A>V | No |
ClinGen gnomAD |
|
|
rs1220541090 CA414974941 |
115 | V>A | No |
ClinGen gnomAD |
|
|
CA10572466 rs781039925 COSM355758 |
115 | V>I | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs1278879967 CA414974960 |
116 | Q>H | No |
ClinGen gnomAD |
|
|
rs1311462417 CA414974975 |
118 | E>* | No |
ClinGen gnomAD |
|
|
CA414974988 rs1208423297 |
118 | E>D | No |
ClinGen gnomAD |
|
|
rs1288137239 CA414974990 |
119 | L>M | No |
ClinGen gnomAD |
|
|
CA414975013 rs1488548781 |
120 | E>D | No |
ClinGen gnomAD |
|
|
rs745768816 CA10572467 |
121 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1240362123 CA414975031 |
122 | V>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs201424738 CA10572468 |
122 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1240362123 CA414975029 |
122 | V>L | No |
ClinGen gnomAD |
|
|
rs1192052574 CA414975056 |
124 | A>P | No |
ClinGen gnomAD |
|
|
CA10572469 rs775446628 |
126 | A>S | No |
ClinGen ExAC |
|
| TCGA novel | 127 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs772185599 CA10572471 |
144 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1556182525 CA414975370 |
147 | R>C | No |
ClinGen Ensembl |
|
|
rs1431837856 CA414975417 |
151 | V>L | No |
ClinGen gnomAD |
|
|
rs1603142738 CA414975690 |
169 | M>L | No |
ClinGen Ensembl |
|
|
CA414976030 rs1297059542 |
190 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 191 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10572482 rs756757652 |
191 | E>A | No |
ClinGen ExAC |
|
|
CA10572483 rs780785122 |
192 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs745321922 CA10572484 |
193 | K>N | No |
ClinGen ExAC |
|
| VAR_016227 | 195 | P>R | No | UniProt | |
|
CA10572485 rs756131625 |
197 | H>N | No |
ClinGen ExAC |
|
|
CA414976142 rs1275117981 |
198 | L>F | No |
ClinGen gnomAD |
|
|
CA10572487 rs749157835 |
199 | C>F | No |
ClinGen ExAC |
|
|
CA414976152 rs1603142783 |
199 | C>R | No |
ClinGen Ensembl |
|
|
rs1366442282 CA414976169 |
200 | K>E | No |
ClinGen gnomAD |
|
|
rs1603142795 CA414976192 |
201 | I>M | No |
ClinGen Ensembl |
|
|
rs771012234 CA10572491 |
206 | R>Q | No |
ClinGen ExAC |
|
|
CA10572490 rs747291345 |
206 | R>W | No |
ClinGen ExAC |
|
|
rs776707915 CA10572492 |
208 | N>S | No |
ClinGen ExAC |
|
|
CA414976352 rs1231837592 |
212 | Q>R | No |
ClinGen gnomAD |
|
| VAR_016228 | 216 | I>F | No | UniProt | |
|
rs1486450497 CA414976436 |
218 | K>R | No |
ClinGen Ensembl |
|
|
rs1215732327 CA414976622 |
229 | A>V | No |
ClinGen gnomAD |
|
|
rs1556182856 CA414976737 |
237 | W>* | No |
ClinGen Ensembl |
|
|
CA414976764 rs1603142842 |
239 | P>L | No |
ClinGen Ensembl |
|
|
CA414976881 rs1453465763 |
247 | R>C | No |
ClinGen gnomAD |
|
|
CA414976891 rs1363870448 |
248 | R>C | No |
ClinGen gnomAD |
|
|
rs1246391705 CA414976972 |
253 | S>N | No |
ClinGen gnomAD |
|
|
rs763277112 CA10572496 |
267 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA414977215 rs1556182928 |
269 | S>F | No |
ClinGen Ensembl |
|
| TCGA novel | 281 | W>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10572498 rs751830669 |
282 | R>C | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 282 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10572499 rs761547400 |
284 | P>T | No |
ClinGen ExAC |
|
|
rs1333134024 CA414977563 |
285 | L>V | No |
ClinGen gnomAD |
|
|
rs767219205 CA10572500 |
287 | Y>S | No |
ClinGen ExAC |
|
|
rs1377437524 CA414977626 |
288 | Y>F | No |
ClinGen gnomAD |
|
|
CA414977720 rs1277739251 |
294 | P>S | No |
ClinGen gnomAD |
|
|
CA414977719 rs1277739251 |
294 | P>T | No |
ClinGen gnomAD |
|
|
CA414977786 rs1358351478 |
299 | E>Q | No |
ClinGen gnomAD |
|
|
CA414977819 rs1228984985 |
300 | T>M | No |
ClinGen gnomAD |
|
|
CA414977830 rs1332965070 |
301 | S>* | No |
ClinGen gnomAD |
|
|
rs1332965070 CA414977834 |
301 | S>L | No |
ClinGen gnomAD |
|
|
rs771192571 CA10572508 |
304 | S>F | No |
ClinGen ExAC gnomAD |
1 associated diseases with Q01534
Without disease ID
5 regional properties for Q01534
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 355 - 632 | IPR000719 |
| domain | Legume lectin domain | 26 - 278 | IPR001220 |
| active_site | Serine/threonine-protein kinase, active site | 476 - 488 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 361 - 384 | IPR017441 |
| binding_site | Legume lectin, beta chain, Mn/Ca-binding site | 142 - 148 | IPR019825 |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| histone binding | Binding to a histone, any of a group of water-soluble proteins found in association with the DNA of eukaryotic or archaeal chromosomes. They are involved in the condensation and coiling of chromosomes during cell division and have also been implicated in gene regulation and DNA replication. They may be chemically modified (methylated, acetlyated and others) to regulate gene transcription. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| gonadal mesoderm development | The process whose specific outcome is the progression of the gonadal mesoderm over time, from its formation to the mature structure. The gonadal mesoderm is the middle layer of the three primary germ layers of the embryo which will go on to form the gonads of the organism. |
| nucleosome assembly | The aggregation, arrangement and bonding together of a nucleosome, the beadlike structural units of eukaryotic chromatin composed of histones and DNA. |
| sex differentiation | The establishment of the sex of an organism by physical differentiation. |
| spermatogenesis | The developmental process by which male germ line stem cells self renew or give rise to successive cell types resulting in the development of a spermatozoa. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O19110 | TSPY1 | Testis-specific Y-encoded protein 1 | Bos taurus (Bovine) | PR |
| Q99457 | NAP1L3 | Nucleosome assembly protein 1-like 3 | Homo sapiens (Human) | PR |
| P55209 | NAP1L1 | Nucleosome assembly protein 1-like 1 | Homo sapiens (Human) | PR |
| P0CV99 | TSPY4 | Testis-specific Y-encoded protein 4 | Homo sapiens (Human) | PR |
| P0CV98 | TSPY3 | Testis-specific Y-encoded protein 3 | Homo sapiens (Human) | PR |
| P0CW00 | TSPY8 | Testis-specific Y-encoded protein 8 | Homo sapiens (Human) | PR |
| P0CW01 | TSPY10 | Testis-specific Y-encoded protein 10 | Homo sapiens (Human) | PR |
| A6NKD2 | TSPY2 | Testis-specific Y-encoded protein 2 | Homo sapiens (Human) | PR |
| Q8N831 | TSPYL6 | Testis-specific Y-encoded-like protein 6 | Homo sapiens (Human) | PR |
| Q9H2G4 | TSPYL2 | Testis-specific Y-encoded-like protein 2 | Homo sapiens (Human) | PR |
| P0DME0 | SETSIP | Protein SETSIP | Homo sapiens (Human) | SS |
| Q01105 | SET | Protein SET | Homo sapiens (Human) | SS |
| Q9H489 | TSPY26P | Putative testis-specific Y-encoded-like protein 3 | Homo sapiens (Human) | PR |
| Q9EQU5 | Set | Protein SET | Mus musculus (Mouse) | EV |
| Q9R1M3 | Tspy1 | Testis-specific Y-encoded protein 1 | Rattus norvegicus (Rat) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRPEGSLTYR | VPERLRQGFC | GVGRAAQALV | CASAKEGTAF | RMEAVQEGAA | GVESEQAALG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EEAVLLLDDI | MAEVEVVAEE | EGLVERREEA | QRAQQAVPGP | GPMTPESAPE | ELLAVQVELE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PVNAQARKAF | SRQREKMERR | RKPHLDRRGA | VIQSVPGFWA | NVIANHPQMS | ALITDEDEDM |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSYMVSLEVG | EEKHPVHLCK | IMLFFRSNPY | FQNKVITKEY | LVNITEYRAS | HSTPIEWYPD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YEVEAYRRRH | HNSSLNFFNW | FSDHNFAGSN | KIAEILCKDL | WRNPLQYYKR | MKPPEEGTET |
| SGDSQLLS |