Q00944
Gene name |
PTK2 (FAK, FAK1) |
Protein name |
Focal adhesion kinase 1 |
Names |
FADK 1, Focal adhesion kinase-related nonkinase, FRNK, p41/p43FRNK, Protein-tyrosine kinase 2, p125FAK, pp125FAK |
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
gga:396416 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
FERM AND PDZ DOMAIN-CONTAINING PROTEIN FAMILY MEMBER (PTHR46221) |
Descriptions
Focal adhesion kinase (FAK) integrates signals from integrin and growth factor receptors to regulate cellular responses including cell adhesion, migration and survival.
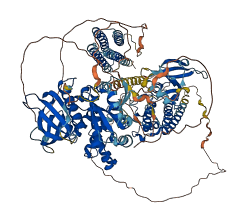
The inactive structure reveals a mechanism of inhibition in which the N-terminal FERM domain directly binds the kinase domain, blocking access to the catalytic cleft and protecting the FAK activation loop from Src phosphorylation. Additionally, the FERM domain sequesters the Tyr397 autophosphorylation and Src recruitment site, which lies in the linker connecting the FERM and kinase domains. FAK is activated by autophosphorylation at Tyr-397, which promotes interaction with Src and phosphorylation at Tyr-576 and Tyr-577 in the kinase activation loop.
Autoinhibitory domains (AIDs)
Target domain |
422-680 (Protein kinase domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
Deletion assay, Mutagenesis experiment, Structural analysis |
Accessory elements
563-587 (Activation loop from InterPro)
Target domain |
422-680 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Lietha D et al. (2007) "Structural basis for the autoinhibition of focal adhesion kinase", Cell, 129, 1177-87
- Choi I et al. (2016) "LRRK2 Inhibits FAK Activity by Promoting FERM-mediated Autoinhibition of FAK and Recruiting the Tyrosine Phosphatase, SHP-2", Experimental neurobiology, 25, 269-276
- Cai X et al. (2008) "Spatial and temporal regulation of focal adhesion kinase activity in living cells", Molecular and cellular biology, 28, 201-14
- Herzog FA et al. (2017) "Structural Insights How PIP2 Imposes Preferred Binding Orientations of FAK at Lipid Membranes", The journal of physical chemistry. B, 121, 3523-3535
Autoinhibited structure
Activated structure
35 structures for Q00944
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1KTM | NMR | - | A | 916-1053 | PDB |
| 1PV3 | NMR | - | A | 920-1053 | PDB |
| 1QVX | NMR | - | A | 920-1053 | PDB |
| 2AEH | X-ray | 253 A | A/B | 31-399 | PDB |
| 2AL6 | X-ray | 235 A | A/B | 31-405 | PDB |
| 2J0J | X-ray | 280 A | A | 31-686 | PDB |
| 2J0K | X-ray | 300 A | A/B | 31-686 | PDB |
| 2J0L | X-ray | 230 A | A | 411-686 | PDB |
| 2J0M | X-ray | 280 A | PDB | ||
| 2JKK | X-ray | 200 A | A | 411-686 | PDB |
| 2JKM | X-ray | 231 A | A | 411-686 | PDB |
| 2JKO | X-ray | 165 A | A | 411-686 | PDB |
| 2JKQ | X-ray | 260 A | A | 411-686 | PDB |
| 2L6F | NMR | - | A | 916-1053 | PDB |
| 2L6G | NMR | - | A | 916-1053 | PDB |
| 2L6H | NMR | - | A | 916-1053 | PDB |
| 3ZDT | X-ray | 315 A | A/B | 31-405 | PDB |
| 4BRX | X-ray | 205 A | A | 411-686 | PDB |
| 4C7T | X-ray | 205 A | A | 411-686 | PDB |
| 4CYE | X-ray | 320 A | A/B | 31-405 | PDB |
| 4D4R | X-ray | 155 A | A/B | 411-686 | PDB |
| 4D4S | X-ray | 200 A | A/B | 411-686 | PDB |
| 4D4V | X-ray | 210 A | A/B | 411-686 | PDB |
| 4D4Y | X-ray | 180 A | A/B | 411-686 | PDB |
| 4D55 | X-ray | 230 A | A | 411-686 | PDB |
| 4D58 | X-ray | 195 A | A/B | 411-686 | PDB |
| 4D5H | X-ray | 175 A | A/B | 411-686 | PDB |
| 4D5K | X-ray | 175 A | A/B | 411-686 | PDB |
| 6CB0 | X-ray | 197 A | A/B | 31-405 | PDB |
| 6GCR | X-ray | 230 A | A | 411-686 | PDB |
| 6GCW | X-ray | 200 A | A/B | 411-686 | PDB |
| 6GCX | X-ray | 155 A | A | 411-686 | PDB |
| 6TY3 | EM | 632 A | A/B | 30-686 | PDB |
| 6TY4 | EM | 596 A | A/B | 30-686 | PDB |
| AF-Q00944-F1 | Predicted | AlphaFoldDB |
10 variants for Q00944
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs731758621 | 106 | N>H | No | Ensembl | |
| rs733675923 | 564 | D>G | No | Ensembl | |
| rs738409887 | 643 | M>I | No | Ensembl | |
| rs739787424 | 647 | C>R | No | Ensembl | |
| rs740816587 | 888 | S>R | No | Ensembl | |
| rs734685984 | 889 | L>P | No | Ensembl | |
| rs740517256 | 905 | I>N | No | Ensembl | |
| rs737460072 | 962 | L>R | No | Ensembl | |
| rs739207746 | 1030 | V>G | No | Ensembl | |
| rs738557295 | 1037 | D>G | No | Ensembl |
No associated diseases with Q00944
11 regional properties for Q00944
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | FERM domain | 35 - 355 | IPR000299 |
| domain | Protein kinase domain | 422 - 680 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 422 - 676 | IPR001245 |
| domain | Focal adhesion kinase, targeting (FAT) domain | 915 - 1046 | IPR005189 |
| active_site | Tyrosine-protein kinase, active site | 542 - 554 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 428 - 454 | IPR017441 |
| domain | FERM central domain | 135 - 250 | IPR019748 |
| domain | Band 4.1 domain | 31 - 258 | IPR019749 |
| domain | Tyrosine-protein kinase, catalytic domain | 422 - 676 | IPR020635 |
| domain | Focal adhesion kinase, N-terminal | 35 - 130 | IPR041390 |
| domain | FAK1/PYK2, FERM domain C-lobe | 254 - 364 | IPR041784 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR46221 | FERM AND PDZ DOMAIN-CONTAINING PROTEIN FAMILY MEMBER |
| PANTHER Subfamily | PTHR46221:SF8 | NON-SPECIFIC PROTEIN-TYROSINE KINASE |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
8 GO annotations of cellular component
| Name | Definition |
|---|---|
| ciliary basal body | A membrane-tethered, short cylindrical array of microtubules and associated proteins found at the base of a eukaryotic cilium (also called flagellum) that is similar in structure to a centriole and derives from it. The cilium basal body is the site of assembly and remodelling of the cilium and serves as a nucleation site for axoneme growth. As well as anchoring the cilium, it is thought to provide a selective gateway regulating the entry of ciliary proteins and vesicles by intraflagellar transport. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| dendritic spine | A small, membranous protrusion from a dendrite that forms a postsynaptic compartment, typically receiving input from a single presynapse. They function as partially isolated biochemical and an electrical compartments. Spine morphology is variable:they can be thin, stubby, mushroom, or branched, with a continuum of intermediate morphologies. They typically terminate in a bulb shape, linked to the dendritic shaft by a restriction. Spine remodeling is though to be involved in synaptic plasticity. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| sarcolemma | The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers. |
9 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| calcium-dependent cysteine-type endopeptidase activity | Catalysis of the hydrolysis of nonterminal peptide bonds in a polypeptide chain by a mechanism using a cysteine residue at the enzyme active center, and requiring the presence of calcium. |
| identical protein binding | Binding to an identical protein or proteins. |
| integrin binding | Binding to an integrin. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| protease binding | Binding to a protease or a peptidase. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
24 GO annotations of biological process
| Name | Definition |
|---|---|
| actin filament organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments. Includes processes that control the spatial distribution of actin filaments, such as organizing filaments into meshworks, bundles, or other structures, as by cross-linking. |
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| angiogenesis involved in wound healing | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels and contribute to the series of events that restore integrity to a damaged tissue, following an injury. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| epidermal growth factor receptor signaling pathway | The series of molecular signals initiated by binding of a ligand to the tyrosine kinase receptor EGFR (ERBB1) on the surface of a cell. The pathway ends with regulation of a downstream cellular process, e.g. transcription. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| negative regulation of anoikis | Any process that stops, prevents or reduces the frequency, rate or extent of anoikis. |
| negative regulation of cell-substrate adhesion | Any process that decreases the frequency, rate or extent of cell-substrate adhesion. Cell-substrate adhesion is the attachment of a cell to the underlying substrate via adhesion molecules. |
| negative regulation of protein autophosphorylation | Any process that stops, prevents or decreases the rate of the phosphorylation by a protein of one or more of its own residues. |
| positive regulation of cell migration | Any process that activates or increases the frequency, rate or extent of cell migration. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of focal adhesion assembly | Any process that activates or increases the frequency, rate or extent of focal adhesion assembly, the establishment and maturation of focal adhesions. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of protein tyrosine kinase activity | Any process that increases the rate, frequency, or extent of protein tyrosine kinase activity. |
| positive regulation of substrate-dependent cell migration, cell attachment to substrate | Any process that activates or increases the frequency, rate or extent of substrate-dependent cell migration, cell attachment to substrate. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| radial glia-guided pyramidal neuron migration | The radial migration of a pyramidal neuron along radial glial cells. |
| regulation of cell adhesion | Any process that modulates the frequency, rate or extent of attachment of a cell to another cell or to the extracellular matrix. |
| response to muscle stretch | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a myofibril being extended beyond its slack length. |
| response to pH | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a pH stimulus. pH is a measure of the acidity or basicity of an aqueous solution. |
| signal complex assembly | The aggregation, arrangement and bonding together of a set of components to form a complex capable of relaying a signal within a cell. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| wound healing, spreading of cells | The migration of a cell along or through a wound gap that contributes to the reestablishment of a continuous surface. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| F1N9Y5 | SYK | Tyrosine-protein kinase SYK | Gallus gallus (Chicken) | SS |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| P00523 | SRC | Proto-oncogene tyrosine-protein kinase Src | Gallus gallus (Chicken) | EV |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| Q14289 | PTK2B | Protein-tyrosine kinase 2-beta | Homo sapiens (Human) | PR |
| Q05397 | PTK2 | Focal adhesion kinase 1 | Homo sapiens (Human) | EV |
| Q9QVP9 | Ptk2b | Protein-tyrosine kinase 2-beta | Mus musculus (Mouse) | PR |
| P34152 | Ptk2 | Focal adhesion kinase 1 | Mus musculus (Mouse) | SS |
| P70600 | Ptk2b | Protein-tyrosine kinase 2-beta | Rattus norvegicus (Rat) | PR |
| O35346 | Ptk2 | Focal adhesion kinase 1 | Rattus norvegicus (Rat) | SS |
| Q95YD4 | kin-32 | Inactive tyrosine-protein kinase kin-32 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAAYLDPNL | NHTPSSSAKT | HLGTGMERSP | GAMERVLKVF | HYFENSSEPT | TWASIIRHGD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ATDVRGIIQK | IVDCHKVKNV | ACYGLRLSHL | QSEEVHWLHL | DMGVSNVREK | FELAHPPEEW |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KYELRIRYLP | KGFLNQFTED | KPTLNFFYQQ | VKNDYMLEIA | DQVDQEIALK | LGCLEIRRSY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GEMRGNALEK | KSNYEVLEKD | VGLRRFFPKS | LLDSVKAKTL | RKLIQQTFRQ | FANLNREESI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LKFFEILSPV | YRFDKECFKC | ALGSSWIISV | ELAIGPEEGI | SYLTDKGANP | THLADFNQVQ |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TIQYSNSEDK | DRKGMLQLKI | AGAPEPLTVT | APSLTIAENM | ADLIDGYCRL | VNGATQSFII |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RPQKEGERAL | PSIPKLANNE | KQGVRSHTVS | VSETDDYAEI | IDEEDTYTMP | STRDYEIQRE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RIELGRCIGE | GQFGDVHQGI | YMSPENPAMA | VAIKTCKNCT | SDSVREKFLQ | EALTMRQFDH |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PHIVKLIGVI | TENPVWIIME | LCTLGELRSF | LQVRKFSLDL | ASLILYAYQL | STALAYLESK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RFVHRDIAAR | NVLVSATDCV | KLGDFGLSRY | MEDSTYYKAS | KGKLPIKWMA | PESINFRRFT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| SASDVWMFGV | CMWEILMHGV | KPFQGVKNND | VIGRIENGER | LPMPPNCPPT | LYSLMTKCWA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| YDPSRRPRFT | ELKAQLSTIL | EEEKLQQEER | MRMESRRQVT | VSWDSGGSDE | APPKPSRPGY |
| 730 | 740 | 750 | 760 | 770 | 780 |
| PSPRSSEGFY | PSPQHMVQPN | HYQVSGYSGS | HGIPAMAGSI | YPGQASLLDQ | TDSWNHRPQE |
| 790 | 800 | 810 | 820 | 830 | 840 |
| VSAWQPNMED | SGTLDVRGMG | QVLPTHLMEE | RLIRQQQEME | EDQRWLEKEE | RFLVMKPDVR |
| 850 | 860 | 870 | 880 | 890 | 900 |
| LSRGSIERED | GGLQGPAGNQ | HIYQPVGKPD | HAAPPKKPPR | PGAPHLGSLA | SLNSPVDSYN |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EGVKIKPQEI | SPPPTANLDR | SNDKVYENVT | GLVKAVIEMS | SKIQPAPPEE | YVPMVKEVGL |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| ALRTLLATVD | ESLPVLPAST | HREIEMAQKL | LNSDLAELIN | KMKLAQQYVM | TSLQQEYKKQ |
| 1030 | 1040 | 1050 | |||
| MLTAAHALAV | DAKNLLDVID | QARLKMISQS | RPH |