Q00013
Gene name |
MPP1 (DXS552E, EMP55) |
Protein name |
55 kDa erythrocyte membrane protein |
Names |
p55, Membrane protein, palmitoylated 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4354 |
EC number |
|
Protein Class |
|
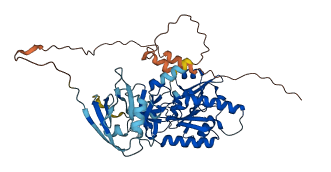
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for Q00013
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2EJY | NMR | - | A | 69-153 | PDB |
| 2EV8 | NMR | - | A | 69-153 | PDB |
| 3NEY | X-ray | 226 A | A/B/C/D/E/F | 282-460 | PDB |
| AF-Q00013-F1 | Predicted | AlphaFoldDB |
219 variants for Q00013
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1557269070 CA414889285 |
2 | T>A | No |
ClinGen gnomAD |
|
|
CA414889272 rs1557269069 |
4 | K>E | No |
ClinGen gnomAD |
|
|
rs1557269065 CA414889245 |
7 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 7 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs984119579 CA337309030 |
8 | G>D | No |
ClinGen Ensembl |
|
|
CA10567685 rs782578742 |
9 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414889235 rs1569559056 |
9 | E>G | No |
ClinGen Ensembl |
|
|
rs1415779741 CA414889225 |
10 | S>R | No |
ClinGen TOPMed |
|
|
rs1426797469 CA414889228 |
10 | S>T | No |
ClinGen TOPMed |
|
|
rs150870475 CA10567684 |
13 | S>G | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs141530265 CA10567683 |
16 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA337309014 rs906703140 |
20 | D>G | No |
ClinGen TOPMed |
|
|
rs1557269052 CA414889155 |
21 | L>H | No |
ClinGen gnomAD |
|
|
rs781863589 CA10567680 |
22 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA337308970 rs1025124382 |
24 | E>K | No |
ClinGen Ensembl |
|
|
rs1557269048 CA414889131 |
25 | H>N | No |
ClinGen gnomAD |
|
|
CA337308949 rs1012422445 |
32 | R>G | No |
ClinGen Ensembl |
|
|
rs782509606 CA10567644 |
35 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA10567645 rs782596654 |
35 | A>T | No |
ClinGen ExAC |
|
|
CA414917456 rs1391055476 |
36 | V>I | No |
ClinGen TOPMed |
|
|
rs781808389 CA10567640 |
37 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 39 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414917336 rs1557267849 |
42 | T>I | No |
ClinGen TOPMed |
|
|
CA414917261 rs1357504901 |
45 | E>D | No |
ClinGen TOPMed |
|
|
CA414917281 rs1557267846 |
45 | E>K | No |
ClinGen gnomAD |
|
|
CA414917228 rs1450368610 |
47 | M>L | No |
ClinGen TOPMed |
|
|
rs1285281979 CA414917217 |
47 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA414917158 rs1557267842 |
49 | T>I | No |
ClinGen gnomAD |
|
|
rs782318569 CA10567638 |
51 | G>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA414917131 rs1569558925 |
51 | G>R | No |
ClinGen Ensembl |
|
|
CA414917074 rs1557267835 |
53 | P>S | No |
ClinGen gnomAD |
|
|
rs1290626416 CA414917039 |
55 | P>Q | No |
ClinGen TOPMed |
|
|
rs782785939 CA10567636 |
56 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA337322044 rs930578863 |
61 | V>A | No |
ClinGen Ensembl |
|
|
RCV000901882 CA10567635 rs143824239 |
61 | V>F | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1557267819 CA414916794 |
66 | V>M | No |
ClinGen gnomAD |
|
|
CA10567633 rs371958769 |
67 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782041042 CA10567634 |
67 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA337322039 rs138328428 |
68 | K>E | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA414916758 rs138328428 |
68 | K>Q | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA414916685 rs1210266367 |
70 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA414916643 rs1557267813 |
72 | I>T | No |
ClinGen gnomAD |
|
|
rs1557267812 CA414916508 |
78 | T>R | No |
ClinGen gnomAD |
|
|
rs1482050291 CA414916435 |
82 | M>L | No |
ClinGen TOPMed |
|
|
CA414916428 rs1184019749 |
82 | M>T | No |
ClinGen TOPMed |
|
|
rs1557267778 CA414916283 |
85 | T>M | No |
ClinGen gnomAD |
|
| TCGA novel | 87 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557267772 CA414916214 |
89 | N>K | No |
ClinGen gnomAD |
|
|
CA10567619 rs782524498 |
95 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA337321885 rs374791319 |
97 | A>S | No |
ClinGen ESP |
|
|
rs374791319 CA337321878 |
97 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP NCI-TCGA |
|
CA414916025 rs1190635283 |
98 | R>K | No |
ClinGen TOPMed |
|
|
CA414915764 rs1557267756 |
107 | R>K | No |
ClinGen gnomAD |
|
|
rs782194140 CA414915583 |
110 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA414915580 rs782194140 |
110 | S>F | No |
ClinGen ExAC gnomAD |
|
|
rs782194140 CA10567606 |
110 | S>Y | No |
ClinGen ExAC gnomAD |
|
|
CA10567605 rs782591996 |
111 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs1557267693 CA414915487 |
115 | D>G | No |
ClinGen gnomAD |
|
|
rs1557267686 CA414915340 |
120 | I>M | No |
ClinGen gnomAD |
|
|
CA10567603 COSM167337 rs782242886 |
121 | N>S | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA414915275 rs1282582153 |
123 | T>A | No |
ClinGen TOPMed |
|
|
rs782646744 CA10567602 |
128 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337321679 rs898856762 |
131 | D>G | No |
ClinGen Ensembl |
|
|
rs148881397 CA10567601 |
136 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1557267575 CA414913338 |
139 | E>A | No |
ClinGen gnomAD |
|
|
CA337321284 rs963538895 |
140 | T>I | No |
ClinGen Ensembl |
|
|
CA414913307 rs1229526197 |
141 | K>R | No |
ClinGen TOPMed |
|
|
rs1270757340 CA414913268 |
143 | M>I | No |
ClinGen TOPMed |
|
|
rs1355347891 CA414913262 |
144 | I>V | No |
ClinGen TOPMed |
|
|
rs1207882398 CA414913238 |
145 | S>L | No |
ClinGen TOPMed |
|
|
rs782274702 CA10567584 |
146 | L>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782029204 CA10567583 |
148 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs782304312 CA10567582 |
149 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs782221902 CA10567581 |
150 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs199536888 CA10567579 |
154 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414913102 rs1247490503 |
155 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs782258559 CA10567578 |
155 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs782678603 CA10567577 |
156 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA414913080 rs1195942350 |
157 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA10567576 rs782555745 |
159 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA414912706 rs1557267200 |
161 | M>I | No |
ClinGen gnomAD |
|
|
rs781929415 CA10567565 |
162 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs929826844 CA337319941 |
163 | M>T | No |
ClinGen Ensembl |
|
|
rs369273483 CA10567564 |
165 | A>T | No |
ClinGen ESP ExAC gnomAD |
|
|
CA414912643 rs1557267196 |
165 | A>V | No |
ClinGen gnomAD |
|
|
CA10567562 rs782006929 |
172 | K>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1467207549 CA414912536 |
173 | K>E | No |
ClinGen TOPMed |
|
|
rs781862397 CA337319889 |
174 | D>G | No |
ClinGen Ensembl |
|
|
CA10567560 rs375730305 |
176 | L>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414912480 rs1557267194 |
178 | P>L | No |
ClinGen gnomAD |
|
| TCGA novel | 180 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM271397 CA10567558 rs782624566 |
182 | A>V | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 183 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414912357 rs1557267191 |
184 | L>R | No |
ClinGen gnomAD |
|
|
rs1399814283 CA414912291 |
186 | F>L | No |
ClinGen TOPMed |
|
|
CA337319819 rs950125120 |
187 | A>T | No |
ClinGen Ensembl |
|
|
rs1557267189 CA414912222 |
189 | G>E | No |
ClinGen gnomAD |
|
|
CA337319798 rs915949014 |
191 | I>V | No |
ClinGen Ensembl |
|
|
rs1342003738 CA414912157 |
192 | I>L | No |
ClinGen TOPMed |
|
|
rs373912523 CA10567552 |
198 | D>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs781784092 CA10567551 |
201 | N>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 206 | R>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782108523 CA10567549 |
206 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10567550 rs782521600 |
206 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs35003354 CA10567547 |
210 | S>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA10567546 rs146452529 |
214 | S>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782034340 CA10567545 |
215 | A>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 216 | G>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs267606401 CA337319739 |
220 | S>F | No |
ClinGen Ensembl |
|
|
rs1187384336 CA414911378 |
221 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1413132238 CA414911288 |
225 | E>K | No |
ClinGen TOPMed |
|
|
CA10567519 rs144066552 |
228 | V>M | No |
ClinGen ESP ExAC TOPMed |
|
|
CA414910884 rs1557267082 |
231 | M>V | No |
ClinGen gnomAD |
|
|
rs782389064 CA10567517 |
235 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs942881402 CA337319274 |
235 | A>V | No |
ClinGen Ensembl |
|
|
rs1557267078 CA414910777 |
236 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 237 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs146674414 CA10567515 |
238 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA414910648 rs1557267075 |
240 | P>A | No |
ClinGen gnomAD |
|
|
CA10567514 rs782301128 |
240 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 247 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10567510 rs782265055 |
248 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA414910338 rs1557267066 |
250 | K>N | No |
ClinGen gnomAD |
|
|
rs143193027 CA10567508 |
253 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782184376 CA10567506 |
258 | K>R | No |
ClinGen ExAC |
|
|
CA10567477 rs781833007 |
263 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs782799952 CA10567476 |
265 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10567475 rs782152401 |
268 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA10567474 rs782190220 |
269 | V>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA414909861 rs1569558819 |
272 | E>K | No |
ClinGen Ensembl |
|
|
rs201515783 CA414909781 |
275 | V>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs201515783 CA10567471 |
275 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782247159 CA10567469 |
276 | R>Q | No |
ClinGen ExAC TOPMed |
|
|
CA10567470 rs782360964 |
276 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA10567468 rs781998412 |
279 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs1603429926 CA414909697 |
280 | F>L | No |
ClinGen Ensembl |
|
|
CA337318899 rs782376251 |
281 | K>E | No |
ClinGen Ensembl |
|
|
rs782417469 CA10567467 |
281 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782285999 CA10567466 |
288 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs149812907 CA10567454 |
291 | S>N | No |
ClinGen ESP ExAC TOPMed |
|
|
rs1480817030 CA414909425 |
291 | S>R | No |
ClinGen TOPMed |
|
|
CA10567453 rs781860060 |
292 | G>E | No |
ClinGen ExAC gnomAD |
|
|
CA10567452 rs782778158 COSM1118171 |
295 | R>C | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs782150002 CA10567451 |
302 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA10567450 rs373372984 |
304 | S>N | No |
ClinGen ESP ExAC TOPMed |
|
|
CA414909155 rs1557266855 |
306 | N>D | No |
ClinGen gnomAD |
|
|
CA10567449 rs782304723 |
306 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10567448 rs782061318 |
307 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs370054543 CA10567447 |
308 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414909066 rs1557266846 |
309 | K>T | No |
ClinGen gnomAD |
|
|
CA414909021 rs1402699708 |
311 | V>L | No |
ClinGen TOPMed |
|
|
rs782048808 CA10567434 |
319 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782671572 CA10567433 |
320 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1557266678 CA414907314 |
332 | H>Y | No |
ClinGen gnomAD |
|
|
CA414907214 rs1557266677 |
335 | S>L | No |
ClinGen gnomAD |
|
|
COSM1466892 rs373323050 CA10567429 |
336 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs201827720 CA10567427 |
340 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs1205013756 CA414906846 |
346 | N>T | No |
ClinGen TOPMed |
|
|
rs1557266668 CA414906779 |
348 | F>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1557266667 CA414906764 |
349 | L>F | No |
ClinGen gnomAD |
|
| TCGA novel | 350 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA414906630 rs1603429861 |
355 | Q>R | No |
ClinGen Ensembl |
|
|
rs781970924 CA10567424 |
357 | N>T | No |
ClinGen ExAC |
|
|
rs1557266663 CA414906557 |
358 | M>L | No |
ClinGen gnomAD |
|
| TCGA novel | 360 | G>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557266659 CA414906432 |
362 | K>R | No |
ClinGen gnomAD |
|
|
rs782382533 CA10567423 |
365 | T>R | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 366 | V>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 366 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA10567420 rs782551298 COSM1190658 |
375 | I>T | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA414905950 rs1557266654 |
377 | I>V | No |
ClinGen gnomAD |
|
|
CA10567419 rs782178153 |
380 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782479508 CA10567417 |
382 | P>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs782479508 CA10567418 |
382 | P>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA414905810 rs1385051125 |
382 | P>S | No |
ClinGen TOPMed |
|
|
rs1557266566 CA414905463 |
386 | K>T | No |
ClinGen gnomAD |
|
|
CA414905394 rs1450303673 |
388 | V>F | No |
ClinGen TOPMed |
|
|
rs376739085 CA10567362 |
389 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA414905381 rs1557266565 |
389 | R>W | No |
ClinGen gnomAD |
|
|
rs1036152470 CA337316743 |
391 | A>S | No |
ClinGen Ensembl |
|
|
CA10567361 rs782353463 |
392 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
rs1557266560 CA414905273 COSM379529 |
394 | S>L | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs781995990 CA10567359 |
398 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA414905146 rs1324714705 |
400 | I>T | No |
ClinGen TOPMed |
|
|
rs782396777 CA10567358 |
400 | I>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA10567357 rs782183476 |
404 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs940488723 CA337316732 |
405 | Q>R | No |
ClinGen Ensembl |
|
|
CA414905002 rs868911141 |
406 | G>D | No |
ClinGen Ensembl |
|
|
CA10567356 rs782580819 |
407 | T>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA337316730 rs200110654 |
408 | Q>R | No |
ClinGen 1000Genomes |
|
|
CA414904417 rs1306044661 |
413 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA414904361 rs1557266338 |
415 | L>V | No |
ClinGen gnomAD |
|
|
CA414904300 rs1229701051 |
416 | Q>R | No |
ClinGen TOPMed |
|
| rs1252010732 | 417 | K>missing | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1557266333 CA414904272 |
417 | K>E | No |
ClinGen gnomAD |
|
|
rs1557266332 CA414904213 |
418 | D>Y | No |
ClinGen gnomAD |
|
|
rs782461332 CA10567338 |
421 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA414903998 rs1356238742 |
421 | A>V | No |
ClinGen TOPMed |
|
|
rs782266951 CA10567337 |
423 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10567336 rs782666257 |
423 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs1557266323 CA414903925 |
424 | S>N | No |
ClinGen gnomAD |
|
|
CA414903709 rs1557266319 |
428 | H>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs782426530 CA10567335 |
430 | F>L | No |
ClinGen ExAC gnomAD |
|
|
CA10567333 rs374639579 |
433 | S>P | No |
ClinGen ESP ExAC gnomAD |
|
| TCGA novel | 443 | L>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs781834831 CA10567331 |
444 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA414903184 rs1569558719 |
445 | K>N | No |
ClinGen Ensembl |
|
|
CA337315784 rs1047904812 |
448 | E>A | No |
ClinGen TOPMed |
|
|
VAR_011914 CA337315789 rs14092 |
448 | E>Q | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs782635278 CA10567330 |
449 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA10567329 rs782511750 |
451 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs781886731 CA10567328 |
452 | Q>L | No |
ClinGen ExAC TOPMed |
|
|
CA414902927 rs781886731 COSM612083 |
452 | Q>P | lung Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed |
|
rs1167015554 CA414902875 |
453 | A>E | No |
ClinGen TOPMed gnomAD |
|
|
COSM1118164 rs1167015554 CA414902865 |
453 | A>V | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs782458500 CA10567326 |
455 | S>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs782458500 CA414902772 |
455 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA337315739 rs938377065 |
458 | Q>H | No |
ClinGen Ensembl |
|
|
CA337315727 rs1044348220 |
467 | Y>E | No |
ClinGen gnomAD |
No associated diseases with Q00013
6 regional properties for Q00013
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH3 domain | 158 - 228 | IPR001452 |
| domain | PDZ domain | 71 - 152 | IPR001478 |
| domain | Guanylate kinase-like domain | 282 - 451 | IPR008144 |
| domain | Guanylate kinase/L-type calcium channel beta subunit | 281 - 454 | IPR008145 |
| conserved_site | Guanylate kinase, conserved site | 317 - 334 | IPR020590 |
| domain | MPP1, SH3 domain | 162 - 223 | IPR035475 |
Functions
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell-cell junction | A cell junction that forms a connection between two or more cells of an organism; excludes direct cytoplasmic intercellular bridges, such as ring canals in insects. |
| centriolar satellite | A small (70-100 nm) cytoplasmic granule that contains a number of centrosomal proteins; centriolar satellites traffic toward microtubule minus ends and are enriched near the centrosome. |
| cortical cytoskeleton | The portion of the cytoskeleton that lies just beneath the plasma membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| stereocilium | An actin-based protrusion from the apical surface of auditory and vestibular hair cells and of neuromast cells. These protrusions are supported by a bundle of cross-linked actin filaments (an actin cable), oriented such that the plus (barbed) ends are at the tip of the protrusion, capped by a tip complex which bridges to the plasma. Bundles of stereocilia act as mechanosensory organelles. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| guanylate kinase activity | Catalysis of the reaction: ATP + GMP = ADP + GDP. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| regulation of neutrophil chemotaxis | Any process that modulates the frequency, rate, or extent of neutrophil chemotaxis. Neutrophil chemotaxis is the directed movement of a neutrophil cell, the most numerous polymorphonuclear leukocyte found in the blood, in response to an external stimulus, usually an infection or wounding. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q24210 | CASK | Peripheral plasma membrane protein CASK | Drosophila melanogaster (Fruit fly) | SS |
| O14936 | CASK | Peripheral plasma membrane protein CASK | Homo sapiens (Human) | EV |
| Q9NZW5 | PALS2 | Protein PALS2 | Homo sapiens (Human) | PR |
| Q14168 | MPP2 | MAGUK p55 subfamily member 2 | Homo sapiens (Human) | PR |
| Q96JB8 | MPP4 | MAGUK p55 subfamily member 4 | Homo sapiens (Human) | PR |
| Q9WV34 | Mpp2 | MAGUK p55 subfamily member 2 | Mus musculus (Mouse) | PR |
| O70589 | Cask | Peripheral plasma membrane protein CASK | Mus musculus (Mouse) | SS |
| O88910 | Mpp3 | MAGUK p55 subfamily member 3 | Mus musculus (Mouse) | PR |
| Q6P7F1 | Mpp4 | MAGUK p55 subfamily member 4 | Mus musculus (Mouse) | PR |
| Q8BVD5 | Mpp7 | MAGUK p55 subfamily member 7 | Mus musculus (Mouse) | PR |
| Q9JLB0 | Pals2 | Protein PALS2 | Mus musculus (Mouse) | PR |
| P70290 | Mpp1 | 55 kDa erythrocyte membrane protein | Mus musculus (Mouse) | PR |
| Q9QYH1 | Mpp4 | MAGUK p55 subfamily member 4 | Rattus norvegicus (Rat) | PR |
| P54936 | lin-2 | Protein lin-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTLKASEGES | GGSMHTALSD | LYLEHLLQKR | SRPEAVSHPL | NTVTEDMYTN | GSPAPGSPAQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VKGQEVRKVR | LIQFEKVTEE | PMGITLKLNE | KQSCTVARIL | HGGMIHRQGS | LHVGDEILEI |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NGTNVTNHSV | DQLQKAMKET | KGMISLKVIP | NQQSRLPALQ | MFMRAQFDYD | PKKDNLIPCK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| EAGLKFATGD | IIQIINKDDS | NWWQGRVEGS | SKESAGLIPS | PELQEWRVAS | MAQSAPSEAP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SCSPFGKKKK | YKDKYLAKHS | SIFDQLDVVS | YEEVVRLPAF | KRKTLVLIGA | SGVGRSHIKN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ALLSQNPEKF | VYPVPYTTRP | PRKSEEDGKE | YHFISTEEMT | RNISANEFLE | FGSYQGNMFG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TKFETVHQIH | KQNKIAILDI | EPQTLKIVRT | AELSPFIVFI | APTDQGTQTE | ALQQLQKDSE |
| 430 | 440 | 450 | 460 | ||
| AIRSQYAHYF | DLSLVNNGVD | ETLKKLQEAF | DQACSSPQWV | PVSWVY |