P86449
Gene name |
Trim43c |
Protein name |
Tripartite motif-containing protein 43C |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:666731 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
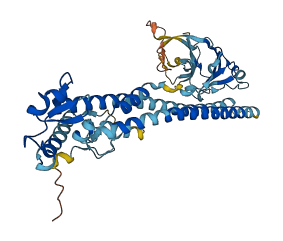
Autoinhibited structure

Activated structure

1 structures for P86449
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P86449-F1 | Predicted | AlphaFoldDB |
32 variants for P86449
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs581430973 | 38 | T>I | No | EVA | |
| rs232794490 | 61 | N>D | No | EVA | |
| rs259443453 | 63 | R>G | No | EVA | |
| rs214015710 | 66 | N>K | No | EVA | |
| rs236887377 | 75 | F>S | No | EVA | |
| rs583354886 | 79 | K>E | No | EVA | |
| rs3400609885 | 92 | K>T | No | EVA | |
| rs580605834 | 128 | S>P | No | EVA | |
| rs3389029736 | 168 | W>* | No | EVA | |
| rs3389060028 | 188 | P>L | No | EVA | |
| rs216573694 | 188 | P>T | No | EVA | |
| rs237157968 | 231 | Q>R | No | EVA | |
| rs3389009150 | 262 | V>M | No | EVA | |
| rs3389064267 | 263 | D>N | No | EVA | |
| rs3389064348 | 283 | R>K | No | EVA | |
| rs3389067025 | 287 | F>L | No | EVA | |
| rs584466776 | 302 | N>T | No | EVA | |
| rs46989951 | 322 | K>E | No | EVA | |
| rs579254242 | 325 | R>C | No | EVA | |
| rs582740945 | 348 | R>G | No | EVA | |
| rs585991291 | 348 | R>K | No | EVA | |
| rs578938285 | 349 | R>Q | No | EVA | |
| rs582396629 | 364 | S>R | No | EVA | |
| rs244541974 | 380 | N>K | No | EVA | |
| rs262064329 | 381 | K>E | No | EVA | |
| rs579966050 | 392 | R>G | No | EVA | |
| rs583150364 | 401 | G>S | No | EVA | |
| rs3389060057 | 405 | V>M | No | EVA | |
| rs3389029767 | 421 | K>M | No | EVA | |
| rs579603003 | 424 | L>F | No | EVA | |
| rs3389060272 | 434 | H>Q | No | EVA | |
| rs48308688 | 442 | F>S | No | EVA |
No associated diseases with P86449
9 regional properties for P86449
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | B-box-type zinc finger | 88 - 129 | IPR000315 |
| domain | Zinc finger, RING-type | 16 - 57 | IPR001841 |
| domain | B30.2/SPRY domain | 271 - 446 | IPR001870 |
| domain | Butyrophylin-like, SPRY domain | 320 - 344 | IPR003879-1 |
| domain | Butyrophylin-like, SPRY domain | 350 - 363 | IPR003879-2 |
| domain | Butyrophylin-like, SPRY domain | 394 - 418 | IPR003879-3 |
| domain | Butyrophylin-like, SPRY domain | 425 - 443 | IPR003879-4 |
| conserved_site | Zinc finger, RING-type, conserved site | 31 - 40 | IPR017907 |
| domain | Zinc finger, C3HC4 RING-type | 16 - 56 | IPR018957 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| zinc ion binding | Binding to a zinc ion (Zn). |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
47 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9G4 | TRIM10 | Tripartite motif-containing protein 10 | Bos taurus (Bovine) | PR |
| Q2T9Z0 | TRIM17 | E3 ubiquitin-protein ligase TRIM17 | Bos taurus (Bovine) | PR |
| E1BJS7 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Bos taurus (Bovine) | PR |
| Q7YRV4 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Bos taurus (Bovine) | PR |
| Q1PRL4 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Gallus gallus (Chicken) | PR |
| Q7YR32 | TRIM10 | Tripartite motif-containing protein 10 | Pan troglodytes (Chimpanzee) | PR |
| O15553 | MEFV | Pyrin | Homo sapiens (Human) | SS |
| Q9BTV5 | FSD1 | Fibronectin type III and SPRY domain-containing protein 1 | Homo sapiens (Human) | PR |
| Q9H2S5 | RNF39 | RING finger protein 39 | Homo sapiens (Human) | PR |
| P19474 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | Homo sapiens (Human) | PR |
| Q9UJV3 | MID2 | Probable E3 ubiquitin-protein ligase MID2 | Homo sapiens (Human) | PR |
| P29590 | PML | Protein PML | Homo sapiens (Human) | PR |
| Q9C029 | TRIM7 | E3 ubiquitin-protein ligase TRIM7 | Homo sapiens (Human) | PR |
| Q9NQ86 | TRIM36 | E3 ubiquitin-protein ligase TRIM36 | Homo sapiens (Human) | PR |
| Q86UV6 | TRIM74 | Tripartite motif-containing protein 74 | Homo sapiens (Human) | PR |
| Q9UPQ4 | TRIM35 | E3 ubiquitin-protein ligase TRIM35 | Homo sapiens (Human) | PR |
| Q6ZMU5 | TRIM72 | Tripartite motif-containing protein 72 | Homo sapiens (Human) | PR |
| Q86UV7 | TRIM73 | Tripartite motif-containing protein 73 | Homo sapiens (Human) | PR |
| Q8N9V2 | TRIML1 | Probable E3 ubiquitin-protein ligase TRIML1 | Homo sapiens (Human) | PR |
| Q86XT4 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Homo sapiens (Human) | PR |
| Q5EBN2 | TRIM61 | Putative tripartite motif-containing protein 61 | Homo sapiens (Human) | PR |
| Q9BZY9 | TRIM31 | E3 ubiquitin-protein ligase TRIM31 | Homo sapiens (Human) | PR |
| Q2Q1W2 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | Homo sapiens (Human) | PR |
| Q9BXM9 | FSD1L | FSD1-like protein | Homo sapiens (Human) | PR |
| A6NCK2 | TRIM43B | Tripartite motif-containing protein 43B | Homo sapiens (Human) | PR |
| Q8BZT2 | Sh3rf2 | E3 ubiquitin-protein ligase SH3RF2 | Mus musculus (Mouse) | PR |
| Q60953 | Pml | Protein PML | Mus musculus (Mouse) | PR |
| Q99PQ1 | Trim12a | Tripartite motif-containing protein 12A | Mus musculus (Mouse) | PR |
| Q61510 | Trim25 | E3 ubiquitin/ISG15 ligase TRIM25 | Mus musculus (Mouse) | PR |
| Q1PSW8 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Mus musculus (Mouse) | PR |
| Q1XH17 | Trim72 | Tripartite motif-containing protein 72 | Mus musculus (Mouse) | PR |
| Q3TL54 | Trim43a | Tripartite motif-containing protein 43A | Mus musculus (Mouse) | PR |
| Q7TPM3 | Trim17 | E3 ubiquitin-protein ligase TRIM17 | Mus musculus (Mouse) | PR |
| Q810I2 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Mus musculus (Mouse) | PR |
| Q9JJ26 | Mefv | Pyrin | Mus musculus (Mouse) | SS |
| Q9WUH5 | Trim10 | Tripartite motif-containing protein 10 | Mus musculus (Mouse) | PR |
| O77666 | TRIM26 | Tripartite motif-containing protein 26 | Sus scrofa (Pig) | PR |
| O19085 | TRIM10 | Tripartite motif-containing protein 10 | Sus scrofa (Pig) | PR |
| Q865W2 | TRIM50 | E3 ubiquitin-protein ligase TRIM50 | Sus scrofa (Pig) | PR |
| Q920M2 | Rnf39 | RING finger protein 39 | Rattus norvegicus (Rat) | PR |
| Q9JJ25 | Mefv | Pyrin | Rattus norvegicus (Rat) | SS |
| A0JPQ4 | Trim72 | Tripartite motif-containing protein 72 | Rattus norvegicus (Rat) | PR |
| Q810I1 | Trim50 | E3 ubiquitin-protein ligase TRIM50 | Rattus norvegicus (Rat) | PR |
| D3ZVM4 | Trim71 | E3 ubiquitin-protein ligase TRIM71 | Rattus norvegicus (Rat) | PR |
| F6QEU4 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| Q640S6 | trim72 | Tripartite motif-containing protein 72 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| E7FAM5 | trim71 | E3 ubiquitin-protein ligase TRIM71 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MESDNLQDPQ | EETLTCSICQ | GIFMDPVYLR | CGHKFCETCL | LLFQEDIKFP | AYCPTCRQPC |
| 70 | 80 | 90 | 100 | 110 | 120 |
| NQRYINDISL | KKQVFIVRKK | RLMEYLNSEE | HKCVTHKAKK | MIFCDKSKIL | LCHLCSDSQE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| HSGHTHCSID | VAVQEKMEEL | LKHMDSLWQR | LKIQQNYVEK | ERRMTLWWLK | SVKLREEVIK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RVYGKQCPPL | SEERDQHIEC | LRHQSNTTLE | ELRKSEATIV | HERNQLTEVY | QELMTMSQRP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YQELLVQDLD | DLFRRSKLAA | KVDMPQGMIP | RLRAHSIPGL | TARLNSFRVK | ISFKHSIMFG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YNSVRPFDIR | LLHESTSLDS | AKTHRVSWGK | KSFSRGKYYW | EVDLKDYRRW | TVGVCKDPWL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RGRSYVVTPT | DIFLLECLRN | KDHYILITRI | GREHYIEKPV | GQVGVFLDCE | GGYVSFVDVA |
| 430 | 440 | ||||
| KSSLILSYSP | GTFHCAVRPF | FFAAYT |