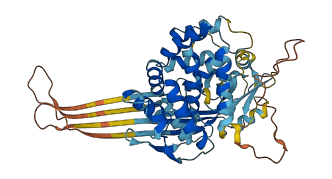
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
4-243 (N-terminal gasdermin domain) |
Relief mechanism |
Cleavage |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P85967
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P85967-F1 | Predicted | AlphaFoldDB |
No variants for P85967
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P85967 | |||||
No associated diseases with P85967
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| phosphatidylinositol-4,5-bisphosphate binding | Binding to phosphatidylinositol-4,5-bisphosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 4' and 5' positions. |
| phosphatidylinositol-4-phosphate binding | Binding to phosphatidylinositol-4-phosphate, a derivative of phosphatidylinositol in which the inositol ring is phosphorylated at the 4' position. |
| phosphatidylserine binding | Binding to phosphatidylserine, a class of glycophospholipids in which a phosphatidyl group is esterified to the hydroxyl group of L-serine. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response to bacterium | Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism. |
| pyroptosis | A caspase-1-dependent cell death subroutine that is associated with the generation of pyrogenic mediators such as IL-1beta and IL-18. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P57764 | GSDMD | Gasdermin-D | Homo sapiens (Human) | EV |
| Q96QA5 | GSDMA | Gasdermin-A | Homo sapiens (Human) | SS |
| Q8TAX9 | GSDMB | Gasdermin-B | Homo sapiens (Human) | EV |
| Q9BYG8 | GSDMC | Gasdermin-C | Homo sapiens (Human) | SS |
| Q32M21 | Gsdma2 | Gasdermin-A2 | Mus musculus (Mouse) | SS |
| Q5Y4Y6 | Gsdma3 | Gasdermin-A3 | Mus musculus (Mouse) | EV |
| Q8CB12 | Gsdmc3 | Gasdermin-C3 | Mus musculus (Mouse) | SS |
| Q9D8T2 | Gsdmd | Gasdermin-D | Mus musculus (Mouse) | SS |
| Q9EST1 | Gsdma | Gasdermin-A | Mus musculus (Mouse) | SS |
| Q3TR54 | Gsdmc4 | Gasdermin-C4 | Mus musculus (Mouse) | SS |
| Q99NB5 | Gsdmc | Gasdermin-C | Mus musculus (Mouse) | SS |
| Q2KHK6 | Gsdmc2 | Gasdermin-C2 | Mus musculus (Mouse) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MLYTFDQVSK | DVVKKLQGKD | LRPVRCLSDA | TKFRQFDILQ | KTPQSLFFKS | EDTPVGYSLL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QILEPNFPVP | ETEVSAPMPL | KHITSQKWKA | DVDVKATIAD | GGASAEFVQS | CGYDIEVQSR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SIPDSKLESL | QNRQGPWGKL | LDKKLSFVTD | CQMGRNNLYV | VTEVFEVTKD | TVVQGSSSID |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LSGKALVSQL | VKGEAQGQWQ | RETTDLVPIP | KGAVLAYKKK | QLVIENNTCA | ILLSANAKKK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| TFPGIFNFGM | SSRSQTMEIV | NSSWIDYIPP | IGRIEEPVHL | DFKYLEKEVF | LRKEQLAMLS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| KDVQDVVFSN | LLPMLSDSDV | LFDLINMLEL | DQLGHMDGPA | GLILDELRKN | SSTPWIDLKG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LILYLLQALM | VLSDTQLDLL | AQSMEMRILL | QQRELVRSIL | EPNFKYPWNI | PFTLQPQLLA |
| 430 | 440 | 450 | 460 | 470 | |
| PLQGEGLAIT | YELLKGCGLK | MEPNSPRSTW | DLEAKMPLSA | LYGILSCLQQ | LVEA |