P84085
Gene name |
ARF5 |
Protein name |
ADP-ribosylation factor 5 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:381 |
EC number |
|
Protein Class |
ADP RIBOSYLATION FACTOR-RELATED (PTHR11711) |
Descriptions
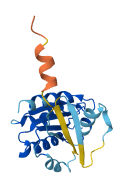
Small GTPases couple their GDP/GTP structural cycle to cytosol/membrane alternation to function as versatile molecular switches in the cell. Membrane localization of their active, GTP-bound form is pivotal to their ability to propagate information, and this requires their post-translational modification by lipids.
Arf GTPases are modified by a myristate attached to their N-terminus, which is shielded by intramolecular interactions in their inactive state. The myristoylated N-terminus of Arf is autoinhibitory in solution and is displaced by membranes, priming Arf GTPases for activation by their GEFs. Replacement of the N-terminal myristate by a 6xHis-tag preserves autoinhibition, representing that membranes unlock the N-terminal region to facilitate subsequent activation.
Autoinhibitory domains (AIDs)
Target domain |
16-138 (Small GTP-binding protein domain) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P84085
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2B6H | X-ray | 176 A | A | 8-180 | PDB |
| AF-P84085-F1 | Predicted | AlphaFoldDB |
121 variants for P84085
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs866219932 | 2 | G>V | No | Ensembl | |
| rs1207747890 | 4 | T>A | No | gnomAD | |
| rs965631736 | 4 | T>I | No |
TOPMed gnomAD |
|
| rs965631736 | 4 | T>S | No |
TOPMed gnomAD |
|
| rs1794237698 | 5 | V>M | No | Ensembl | |
| rs759843060 | 7 | A>V | No |
ExAC gnomAD |
|
| rs1794237834 | 8 | L>F | No | TOPMed | |
| rs1162647178 | 8 | L>P | No | gnomAD | |
| rs1383146791 | 9 | F>I | No | gnomAD | |
| rs1562953554 | 11 | R>P | No | Ensembl | |
| rs1794237934 | 11 | R>W | No | Ensembl | |
| rs769953515 | 12 | I>V | No |
ExAC gnomAD |
|
| rs866714062 | 14 | G>V | No | Ensembl | |
| TCGA novel | 15 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1371360324 | 17 | Q>H | No | gnomAD | |
| rs775454058 | 18 | M>I | No |
ExAC gnomAD |
|
| rs202016109 | 19 | R>L | No | Ensembl | |
| TCGA novel | 19 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1284054298 | 22 | M>I | No | gnomAD | |
| rs1449393189 | 22 | M>L | No | gnomAD | |
| TCGA novel | 24 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1794247263 | 26 | D>A | No | TOPMed | |
| rs866133430 | 27 | A>V | No | Ensembl | |
| rs1794247376 | 28 | A>V | No |
TOPMed gnomAD |
|
| rs1587535775 | 30 | K>Q | No | Ensembl | |
| rs1587535782 | 31 | T>N | No | Ensembl | |
| rs767832041 | 33 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1794247724 | 36 | K>R | No | Ensembl | |
| rs1803150 | 39 | L>W | No | Ensembl | |
| rs1303907072 | 40 | G>E | No |
TOPMed gnomAD |
|
| rs1303907072 | 40 | G>V | No |
TOPMed gnomAD |
|
| rs1587535793 | 41 | E>G | No | TOPMed | |
| rs1794247884 | 41 | E>K | No | gnomAD | |
| COSM1447851 | 41 | E>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 42 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1794247985 | 44 | T>S | No | TOPMed | |
| rs1794248143 | 49 | I>T | No | Ensembl | |
| rs952591454 | 49 | I>V | No | Ensembl | |
| rs912240810 | 51 | F>Y | No |
TOPMed gnomAD |
|
| rs754222019 | 61 | I>T | No | Ensembl | |
| rs1039475305 | 61 | I>V | No | TOPMed | |
| rs1794253997 | 62 | C>R | No | TOPMed | |
| rs1794253997 | 62 | C>S | No | TOPMed | |
| rs981999965 | 74 | I>M | No | Ensembl | |
| rs1297082014 | 75 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM3877865 | 79 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1794254487 | 83 | Q>R | No | Ensembl | |
| rs144009854 | 97 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs367999369 | 97 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| COSM78870 | 98 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
TCGA novel rs1794261468 |
98 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| COSM3698147 | 99 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1794261568 | 101 | Q>* | No | gnomAD | |
| COSM1085148 | 102 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 103 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771445967 | 103 | S>P | No |
ExAC gnomAD |
|
| rs781588986 | 104 | A>T | No |
ExAC gnomAD |
|
| TCGA novel | 108 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 108 | Q>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1389309924 | 110 | M>I | No | gnomAD | |
| rs1794262196 | 110 | M>L | No | Ensembl | |
| rs1794262196 | 110 | M>V | No | Ensembl | |
| rs1223710373 | 114 | D>N | No | gnomAD | |
| rs1489738157 | 115 | E>D | No |
TOPMed gnomAD |
|
|
COSM3674819 rs780593712 |
117 | R>L | prostate [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs780593712 | 117 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs1286936912 COSM1085149 |
117 | R>W | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1176017535 | 118 | D>G | No | TOPMed | |
| rs1233003773 | 119 | A>T | No | gnomAD | |
| rs1794279276 | 119 | A>V | No | TOPMed | |
| rs747749998 | 120 | V>G | No |
ExAC TOPMed gnomAD |
|
| rs1794279479 | 124 | F>C | No | Ensembl | |
|
rs773043755 COSM318745 |
125 | A>S | lung [Cosmic] | No |
cosmic curated ExAC gnomAD |
| TCGA novel | 129 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746526652 | 130 | M>I | No |
ExAC gnomAD |
|
| rs1794279688 | 132 | N>S | No | TOPMed | |
| rs201108741 | 133 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1309906980 | 134 | M>V | No |
TOPMed gnomAD |
|
| TCGA novel | 135 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1794279862 | 135 | P>L | No | TOPMed | |
| rs759398219 | 135 | P>T | No |
ExAC gnomAD |
|
| rs1278238405 | 136 | V>M | No |
TOPMed gnomAD |
|
|
rs775287559 TCGA novel |
137 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA ExAC TOPMed gnomAD |
| rs543068324 | 138 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs543068324 | 138 | E>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1356798060 | 140 | T>S | No | gnomAD | |
| rs1206926592 | 142 | K>R | No | gnomAD | |
| rs1587536937 | 144 | G>A | No | Ensembl | |
| rs1273089709 | 148 | L>F | No | gnomAD | |
| rs1304516534 | 148 | L>S | No | TOPMed | |
| rs751785636 | 149 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs767496095 | 149 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs767496095 | 149 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs751785636 | 149 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs916226462 | 150 | S>G | No |
TOPMed gnomAD |
|
| rs750899407 | 151 | R>C | No |
ExAC gnomAD |
|
| rs756706436 | 151 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs756706436 | 151 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs749643254 | 152 | T>K | No |
ExAC gnomAD |
|
| rs749643254 | 152 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| COSM1085150 | 155 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1222389800 | 157 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1261127308 | 157 | A>V | No | gnomAD | |
| rs780948404 | 158 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs780948404 | 158 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs2117418854 | 162 | Q>P | No | Ensembl | |
| rs1794283006 | 164 | T>K | No |
TOPMed gnomAD |
|
| TCGA novel | 165 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1480770986 | 168 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2117418891 | 171 | D>H | No | Ensembl | |
| COSM744675 | 172 | W>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1794283697 | 174 | S>C | No | TOPMed | |
| rs1794283697 | 174 | S>F | No | TOPMed | |
| rs142827438 | 175 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs774035808 | 176 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1261768055 | 178 | S>* | No |
TOPMed gnomAD |
|
| rs944602969 | 178 | S>A | No |
TOPMed gnomAD |
|
| rs772150653 | 179 | K>N | No |
ExAC gnomAD |
|
|
rs773554378 COSM1085151 |
180 | R>C | ovary Variant assessed as Somatic; MODERATE impact. endometrium [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
COSM1447853 rs760826467 |
180 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1587537120 | 181 | R>Y | No | Ensembl |
No associated diseases with P84085
1 regional properties for P84085
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 16 - 138 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11711 | ADP RIBOSYLATION FACTOR-RELATED |
| PANTHER Subfamily | PTHR11711:SF323 | ADP-RIBOSYLATION FACTOR 5 |
| PANTHER Protein Class | G-protein | |
| PANTHER Pathway Category |
Huntington disease ARF |
|
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | The directed movement of substances from the Golgi back to the endoplasmic reticulum, mediated by vesicles bearing specific protein coats such as COPI or COG. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
48 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P84077 | ARF1 | ADP-ribosylation factor 1 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGLTVSALFS | RIFGKKQMRI | LMVGLDAAGK | TTILYKLKLG | EIVTTIPTIG | FNVETVEYKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ICFTVWDVGG | QDKIRPLWRH | YFQNTQGLIF | VVDSNDRERV | QESADELQKM | LQEDELRDAV |
| 130 | 140 | 150 | 160 | 170 | |
| LLVFANKQDM | PNAMPVSELT | DKLGLQHLRS | RTWYVQATCA | TQGTGLYDGL | DWLSHELSKR |