P84077
Gene name |
ARF1 |
Protein name |
ADP-ribosylation factor 1 |
Names |
EC 3.6.5.2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:375 |
EC number |
3.6.5.2: Acting on GTP; involved in cellular and subcellular movement |
Protein Class |
ADP RIBOSYLATION FACTOR-RELATED (PTHR11711) |
Descriptions
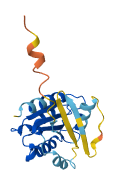
Small GTPases couple their GDP/GTP structural cycle to cytosol/membrane alternation to function as versatile molecular switches in the cell. Membrane localization of their active, GTP-bound form is pivotal to their ability to propagate information, and this requires their post-translational modification by lipids.
Arf GTPases are modified by a myristate attached to their N-terminus, which is shielded by intramolecular interactions in their inactive state. The myristoylated N-terminus of Arf is autoinhibitory in solution and is displaced by membranes, priming Arf GTPases for activation by their GEFs. Replacement of the N-terminal myristate by a 6xHis-tag preserves autoinhibition, representing that membranes unlock the N-terminal region to facilitate subsequent activation.
Autoinhibitory domains (AIDs)
Target domain |
16-141 (Small GTP-binding protein domain) |
Relief mechanism |
Ligand binding |
Assay |
Mutagenesis experiment |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

27 structures for P84077
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1HUR | X-ray | 200 A | A/B | 2-181 | PDB |
| 1RE0 | X-ray | 240 A | A | 18-181 | PDB |
| 1U81 | NMR | - | A | 18-181 | PDB |
| 3O47 | X-ray | 280 A | A/B | 11-181 | PDB |
| 4HMY | X-ray | 700 A | C | 17-181 | PDB |
| 6CM9 | EM | 373 A | C/H | 17-181 | PDB |
| 6CRI | EM | 680 A | C/H/K/L/U/V | 17-181 | PDB |
| 6D83 | EM | 427 A | C/H | 17-181 | PDB |
| 6D84 | EM | 672 A | C/H/I/N | 17-181 | PDB |
| 6DFF | EM | 390 A | C/H | 17-181 | PDB |
| 6FAE | X-ray | 235 A | B | 18-181 | PDB |
| 7DN8 | X-ray | 261 A | B/D/F/H | 17-181 | PDB |
| 7DN9 | X-ray | 329 A | B/D/F/H | 17-181 | PDB |
| 7MGE | EM | 394 A | E | 17-180 | PDB |
| 7R4H | EM | 234 A | C/H | 17-181 | PDB |
| 7UX3 | EM | 960 A | C/H | 2-181 | PDB |
| 8D4C | EM | 930 A | C/D/F/H | 2-181 | PDB |
| 8D4D | EM | 960 A | C/D/F/H | 2-181 | PDB |
| 8D4E | EM | 920 A | C/H/Z | 2-181 | PDB |
| 8D4F | EM | 980 A | C/D/F/H/Q/Z | 2-181 | PDB |
| 8D4G | EM | 1160 A | C/D/F/H/Q/Z | 1-181 | PDB |
| 8D9R | EM | 2000 A | 3/4/5/6/7/C/H/J/K/O/P/Q/X/Z/a/b/c/d | 2-181 | PDB |
| 8D9S | EM | 2000 A | 3/4/5/6/7/C/H/J/K/O/P/Q/X/Z/a/b/c/d | 2-181 | PDB |
| 8D9U | EM | 2000 A | C/H/J/K/O/P/Q/X/Z/a/b/c | 2-181 | PDB |
| 8D9W | EM | 930 A | F/I | 2-181 | PDB |
| 8SDW | X-ray | 175 A | A | 1-181 | PDB |
| AF-P84077-F1 | Predicted | AlphaFoldDB |
87 variants for P84077
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs2124857065 COSM1183326 RCV001845032 |
19 | R>C | Periventricular nodular heterotopia 8 Variant assessed as Somatic; MODERATE impact. large_intestine [ClinVar, NCI-TCGA, Cosmic] | Yes |
NCI-TCGA Cosmic cosmic curated ClinVar Ensembl dbSNP |
|
rs879036238 RCV001845033 |
35 | Y>D | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs879036238 VAR_081272 RCV000721169 |
35 | Y>H | Periventricular nodular heterotopia 8 PVNH8; decreased interaction with GGA3 [ClinVar, UniProt] | Yes |
ClinVar UniProt Ensembl dbSNP |
|
rs1571844539 RCV000851181 |
48 | T>I | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001845034 rs2124857424 |
51 | F>L | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs2124857427 RCV001845029 |
51 | F>L | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs2124857526 RCV001843892 |
72 | D>missing | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2124857932 RCV001845035 |
93 | D>G | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs1571845049 RCV000789499 |
98 | E>A | Charcot-Marie-Tooth disease [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
rs1571845049 RCV000789500 |
98 | E>G | Charcot-Marie-Tooth disease [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001845036 rs2124857939 |
99 | R>C | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001579777 rs1558087712 VAR_081273 RCV000721171 |
99 | R>H | Periventricular nodular heterotopia 8 PVNH8; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
VAR_081274 RCV000721170 rs1558087795 |
127 | K>E | Periventricular nodular heterotopia 8 PVNH8 [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
rs2124858360 RCV001845031 |
131 | P>L | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
|
RCV001845030 rs2124858360 |
131 | P>R | Periventricular nodular heterotopia 8 [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
| rs2124856973 | 3 | N>S | No | Ensembl | |
| COSM3689392 | 4 | I>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2032773362 | 4 | I>V | No | gnomAD | |
| rs201883341 | 6 | A>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs778178441 | 7 | N>S | No |
ExAC gnomAD |
|
| rs747344148 | 10 | K>R | No |
ExAC gnomAD |
|
| TCGA novel | 11 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs11541562 | 12 | L>P | No | Ensembl | |
| TCGA novel | 13 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1448468387 | 15 | K>R | No | gnomAD | |
| TCGA novel | 17 | E>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1339437 | 17 | E>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2124857075 | 22 | M>L | No | Ensembl | |
| COSM904931 | 25 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1571844470 | 31 | T>P | No | Ensembl | |
| rs1434233811 | 32 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM904932 | 36 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2124857181 | 40 | G>D | No | Ensembl | |
| TCGA novel | 43 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1571844523 | 44 | T>P | No | Ensembl | |
| COSM464078 | 61 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1182062930 | 65 | V>A | No | gnomAD | |
|
RCV001311681 rs2032786415 |
66 | W>R | No |
ClinVar Ensembl dbSNP |
|
| rs1476367998 | 75 | R>W | No | gnomAD | |
|
COSM5643928 rs752069371 |
79 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| COSM4028850 | 79 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs752069371 | 79 | R>S | No |
ExAC gnomAD |
|
| rs1457087481 | 82 | F>L | No | gnomAD | |
| rs2032794637 | 88 | L>V | No | TOPMed | |
| rs2032794801 | 89 | I>V | No | TOPMed | |
| COSM4028851 | 91 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1313181955 | 101 | N>S | No | TOPMed | |
|
rs1447268730 COSM4028852 |
104 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
|
COSM4028853 rs901254200 |
108 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
| rs2032795613 | 108 | M>V | No | Ensembl | |
| rs11541556 | 110 | M>I | No | Ensembl | |
| TCGA novel | 112 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752583036 | 117 | R>Q | No |
ExAC gnomAD |
|
| TCGA novel | 119 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs373715936 | 124 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM904934 rs1382592951 |
125 | A>T | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1571845119 | 126 | N>K | No | Ensembl | |
| COSM5182008 | 126 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1157677154 | 133 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2032804281 | 137 | A>S | No | TOPMed | |
|
COSM4391484 rs2032804281 |
137 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs776377227 | 137 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| COSM6124799 | 139 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1480071074 | 146 | H>R | No |
TOPMed gnomAD |
|
| rs1409697252 | 147 | S>L | No | gnomAD | |
|
COSM425499 rs1419831068 |
149 | R>H | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs952976361 | 150 | H>D | No | TOPMed | |
| TCGA novel | 151 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 151 | R>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2032806188 | 155 | I>F | No | TOPMed | |
| rs1571845443 | 158 | T>P | No | Ensembl | |
| TCGA novel | 158 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1444487351 | 159 | C>G | No | gnomAD | |
| rs1415720170 | 162 | S>G | No | gnomAD | |
|
TCGA novel rs2032807541 |
164 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1307521193 | 165 | G>W | No | gnomAD | |
| rs748951714 | 168 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1206052329 | 168 | E>K | No | gnomAD | |
|
RCV002246972 rs2124858573 |
170 | L>Q | No |
ClinVar Ensembl dbSNP |
|
| rs768348405 | 171 | D>E | No |
ExAC gnomAD |
|
| rs2032808332 | 172 | W>* | No | TOPMed | |
| rs773772662 | 174 | S>A | No | ExAC | |
| TCGA novel | 175 | N>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747640292 | 175 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs963764078 | 178 | R>Q | No | Ensembl | |
| rs1571845542 | 179 | N>D | No | Ensembl | |
| rs1571845552 | 182 | K>G | No | Ensembl |
1 associated diseases with P84077
[MIM: 618185]: Periventricular nodular heterotopia 8 (PVNH8)
A form of periventricular nodular heterotopia, a disorder resulting from a defect in the pattern of neuronal migration in which ectopic collections of neurons lie along the lateral ventricles of the brain or just beneath, contiguously or in isolated patches. PVNH8 is an autosomal dominant disease characterized by developmental disabilities, speech delay, seizures and attention deficit hyperactivity disorder. . Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of periventricular nodular heterotopia, a disorder resulting from a defect in the pattern of neuronal migration in which ectopic collections of neurons lie along the lateral ventricles of the brain or just beneath, contiguously or in isolated patches. PVNH8 is an autosomal dominant disease characterized by developmental disabilities, speech delay, seizures and attention deficit hyperactivity disorder. . Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P84077
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 16 - 141 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.5.2 | Acting on GTP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11711 | ADP RIBOSYLATION FACTOR-RELATED |
| PANTHER Subfamily | PTHR11711:SF357 | ADP-RIBOSYLATION FACTOR 1 |
| PANTHER Protein Class | G-protein | |
| PANTHER Pathway Category |
Integrin signalling pathway Arf1 Huntington disease ARF |
|
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell leading edge | The area of a motile cell closest to the direction of movement. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| postsynaptic density | An electron dense network of proteins within and adjacent to the postsynaptic membrane of an asymmetric, neuron-neuron synapse. Its major components include neurotransmitter receptors and the proteins that spatially and functionally organize them such as anchoring and scaffolding molecules, signaling enzymes and cytoskeletal components. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| sarcomere | The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to virus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus. |
| dendritic spine organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a dendritic spine. A dendritic spine is a specialized protrusion from a neuronal dendrite and is involved in synaptic transmission. |
| intracellular copper ion homeostasis | A homeostatic process involved in the maintenance of a steady state level of copper ions within a cell. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
| long-term synaptic depression | A process that modulates synaptic plasticity such that synapses are changed resulting in the decrease in the rate, or frequency of synaptic transmission at the synapse. |
| mitotic cleavage furrow ingression | Advancement of the mitotic cleavage furrow from the outside of the cell inward towards the center of the cell. The cleavage furrow acts as a 'purse string' which draws tight to separate daughter cells during mitotic cytokinesis and partition the cytoplasm between the two daughter cells. The furrow ingresses until a cytoplasmic bridge is formed. |
| regulation of Arp2/3 complex-mediated actin nucleation | Any process that modulates the frequency, rate or extent of actin nucleation mediated by the Arp2/3 complex and interacting proteins. |
| regulation of receptor internalization | Any process that modulates the frequency, rate or extent of receptor internalization. |
| vesicle-mediated transport | A cellular transport process in which transported substances are moved in membrane-bounded vesicles; transported substances are enclosed in the vesicle lumen or located in the vesicle membrane. The process begins with a step that directs a substance to the forming vesicle, and includes vesicle budding and coating. Vesicles are then targeted to, and fuse with, an acceptor membrane. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2KJ96 | ARL5A | ADP-ribosylation factor-like protein 5A | Bos taurus (Bovine) | SS |
| P84081 | ARF2 | ADP-ribosylation factor 2 | Bos taurus (Bovine) | SS |
| Q3SZF2 | ARF4 | ADP-ribosylation factor 4 | Bos taurus (Bovine) | SS |
| Q0VC18 | ARL4D | ADP-ribosylation factor-like protein 4D | Bos taurus (Bovine) | PR |
| P84080 | ARF1 | ADP-ribosylation factor 1 | Bos taurus (Bovine) | SS |
| Q5E9I6 | ARF3 | ADP-ribosylation factor 3 | Bos taurus (Bovine) | SS |
| P49702 | ARF5 | ADP-ribosylation factor 5 | Gallus gallus (Chicken) | SS |
| P40945 | Arf4 | ADP ribosylation factor 4 | Drosophila melanogaster (Fruit fly) | SS |
| P61209 | Arf1 | ADP-ribosylation factor 1 | Drosophila melanogaster (Fruit fly) | SS |
| A6NH57 | ARL5C | Putative ADP-ribosylation factor-like protein 5C | Homo sapiens (Human) | SS |
| P18085 | ARF4 | ADP-ribosylation factor 4 | Homo sapiens (Human) | EV |
| P49703 | ARL4D | ADP-ribosylation factor-like protein 4D | Homo sapiens (Human) | PR |
| P84085 | ARF5 | ADP-ribosylation factor 5 | Homo sapiens (Human) | EV |
| Q8N4G2 | ARL14 | ADP-ribosylation factor-like protein 14 | Homo sapiens (Human) | SS |
| Q969Q4 | ARL11 | ADP-ribosylation factor-like protein 11 | Homo sapiens (Human) | PR |
| Q96KC2 | ARL5B | ADP-ribosylation factor-like protein 5B | Homo sapiens (Human) | SS |
| P62330 | ARF6 | ADP-ribosylation factor 6 | Homo sapiens (Human) | EV |
| Q8IVW1 | ARL17A | ADP-ribosylation factor-like protein 17 | Homo sapiens (Human) | SS |
| Q9H0F7 | ARL6 | ADP-ribosylation factor-like protein 6 | Homo sapiens (Human) | SS |
| P40616 | ARL1 | ADP-ribosylation factor-like protein 1 | Homo sapiens (Human) | SS |
| P61204 | ARF3 | ADP-ribosylation factor 3 | Homo sapiens (Human) | EV |
| Q9Y689 | ARL5A | ADP-ribosylation factor-like protein 5A | Homo sapiens (Human) | SS |
| P49076 | ARF1 | ADP-ribosylation factor | Zea mays (Maize) | SS |
| P84084 | Arf5 | ADP-ribosylation factor 5 | Mus musculus (Mouse) | SS |
| P61750 | Arf4 | ADP-ribosylation factor 4 | Mus musculus (Mouse) | SS |
| Q99PE9 | Arl4d | ADP-ribosylation factor-like protein 4D | Mus musculus (Mouse) | PR |
| Q6P068 | Arl5c | ADP-ribosylation factor-like protein 5C | Mus musculus (Mouse) | SS |
| Q80ZU0 | Arl5a | ADP-ribosylation factor-like protein 5A | Mus musculus (Mouse) | SS |
| P61205 | Arf3 | ADP-ribosylation factor 3 | Mus musculus (Mouse) | SS |
| Q9D4P0 | Arl5b | ADP-ribosylation factor-like protein 5B | Mus musculus (Mouse) | SS |
| Q8BSL7 | Arf2 | ADP-ribosylation factor 2 | Mus musculus (Mouse) | SS |
| P84078 | Arf1 | ADP-ribosylation factor 1 | Mus musculus (Mouse) | SS |
| P51824 | ADP-ribosylation factor 1 | Solanum tuberosum (Potato) | SS | |
| P84082 | Arf2 | ADP-ribosylation factor 2 | Rattus norvegicus (Rat) | SS |
| P51646 | Arl5a | ADP-ribosylation factor-like protein 5A | Rattus norvegicus (Rat) | SS |
| P84083 | Arf5 | ADP-ribosylation factor 5 | Rattus norvegicus (Rat) | SS |
| P36407 | Trim23 | E3 ubiquitin-protein ligase TRIM23 | Rattus norvegicus (Rat) | SS |
| P61751 | Arf4 | ADP-ribosylation factor 4 | Rattus norvegicus (Rat) | SS |
| P61206 | Arf3 | ADP-ribosylation factor 3 | Rattus norvegicus (Rat) | SS |
| P84079 | Arf1 | ADP-ribosylation factor 1 | Rattus norvegicus (Rat) | SS |
| P51823 | ARF | ADP-ribosylation factor 2 | Oryza sativa subsp. japonica (Rice) | SS |
| Q06396 | Os01g0813400 | ADP-ribosylation factor 1 | Oryza sativa subsp. japonica (Rice) | SS |
| P34212 | arl-5 | ADP-ribosylation factor-like protein 5 | Caenorhabditis elegans | SS |
| Q10943 | arf-1.2 | ADP-ribosylation factor 1-like 2 | Caenorhabditis elegans | SS |
| P40940 | ARF3 | ADP-ribosylation factor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P0DH91 | ARF2-B | ADP-ribosylation factor 2-B | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P36397 | ARF1 | ADP-ribosylation factor 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LQC8 | ARF2-A | ADP-ribosylation factor 2-A | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5M9P8 | arl6 | ADP-ribosylation factor-like protein 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGNIFANLFK | GLFGKKEMRI | LMVGLDAAGK | TTILYKLKLG | EIVTTIPTIG | FNVETVEYKN |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ISFTVWDVGG | QDKIRPLWRH | YFQNTQGLIF | VVDSNDRERV | NEAREELMRM | LAEDELRDAV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LLVFANKQDL | PNAMNAAEIT | DKLGLHSLRH | RNWYIQATCA | TSGDGLYEGL | DWLSNQLRNQ |
| K |