P80192
Gene name |
MAP3K9 (MLK1, PRKE1) |
Protein name |
Mitogen-activated protein kinase kinase kinase 9 |
Names |
EC 2.7.11.25 , Mixed lineage kinase 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4293 |
EC number |
2.7.11.-: Protein-serine/threonine kinases |
Protein Class |
|
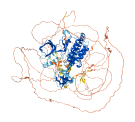
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
490-512 (Proline-rich region) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
293-314 (Activation loop from InterPro)
Target domain |
144-412 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Zhang H et al. (2001) "Autoinhibition of mixed lineage kinase 3 through its Src homology 3 domain", The Journal of biological chemistry, 276, 45598-603
- Du Y et al. (2005) "Cdc42 induces activation loop phosphorylation and membrane targeting of mixed lineage kinase 3", The Journal of biological chemistry, 280, 42984-93
- Handley ME et al. (2007) "Mixed lineage kinases (MLKs): a role in dendritic cells, inflammation and immunity", International journal of experimental pathology, 88, 111-26
- Kokoszka ME et al. (2018) "Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3", The Journal of biological chemistry, 293, 13553-13565
Autoinhibited structure

Activated structure

3 structures for P80192
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3DTC | X-ray | 260 A | A | 136-406 | PDB |
| 4UY9 | X-ray | 281 A | A/B | 135-456 | PDB |
| AF-P80192-F1 | Predicted | AlphaFoldDB |
1188 variants for P80192
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs2055037752 | 3 | P>L | No | gnomAD | |
| rs2055037648 | 5 | R>G | No | gnomAD | |
| rs1272403745 | 6 | A>T | No | TOPMed | |
| rs2139878402 | 9 | G>S | No | Ensembl | |
| rs2055037348 | 10 | C>F | No | TOPMed | |
| rs2055037207 | 14 | A>T | No | Ensembl | |
| rs2055037152 | 14 | A>V | No | Ensembl | |
| rs1282524072 | 16 | A>T | No | gnomAD | |
| rs1014554355 | 18 | A>V | No |
TOPMed gnomAD |
|
| rs1005081428 | 19 | P>L | No | TOPMed | |
| rs887529850 | 20 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs753691995 | 21 | G>W | No |
ExAC TOPMed gnomAD |
|
| rs960014464 | 22 | E>K | No | TOPMed | |
| rs2055036474 | 23 | D>N | No | TOPMed | |
| rs1479118819 | 24 | G>A | No |
TOPMed gnomAD |
|
| rs1173430231 | 25 | A>E | No | gnomAD | |
| rs1034427131 | 26 | G>R | No |
TOPMed gnomAD |
|
| rs1034427131 | 26 | G>W | No |
TOPMed gnomAD |
|
| rs36280 | 27 | A>S | No |
TOPMed gnomAD |
|
| rs36280 | 27 | A>T | No |
TOPMed gnomAD |
|
| rs1427244393 | 27 | A>V | No | gnomAD | |
| rs1473170311 | 28 | G>E | No | gnomAD | |
| rs766214214 | 28 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs2055035982 | 29 | A>V | No | TOPMed | |
| TCGA novel | 30 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1294715594 | 31 | E>G | No | Ensembl | |
| rs1437672891 | 32 | E>K | No | gnomAD | |
| rs1201861960 | 33 | E>G | No |
TOPMed gnomAD |
|
| rs1256394999 | 33 | E>K | No | gnomAD | |
| rs1201861960 | 33 | E>V | No |
TOPMed gnomAD |
|
| rs1001587212 | 36 | E>A | No |
TOPMed gnomAD |
|
| rs1198593145 | 36 | E>K | No | gnomAD | |
| rs1459725624 | 37 | E>A | No | gnomAD | |
| rs2055035345 | 37 | E>K | No | gnomAD | |
|
rs201322413 COSM1214457 |
38 | E>A | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs201322413 | 38 | E>G | No |
ExAC TOPMed gnomAD |
|
| rs1228479489 | 38 | E>K | No | gnomAD | |
|
COSM1214458 rs200801204 |
39 | A>E | large_intestine [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1277763391 | 39 | A>T | No | gnomAD | |
| rs200801204 | 39 | A>V | No |
TOPMed gnomAD |
|
| rs1431812059 | 40 | A>E | No | TOPMed | |
| rs1431812059 | 40 | A>V | No | TOPMed | |
| rs946151572 | 41 | A>V | No | gnomAD | |
| rs199678931 | 42 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1454740532 | 42 | A>T | No | gnomAD | |
| rs199678931 | 42 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1387274267 | 43 | V>L | No | TOPMed | |
| rs1365322420 | 44 | G>D | No | TOPMed | |
| rs1313052429 | 45 | P>L | No |
TOPMed gnomAD |
|
| rs2055033760 | 45 | P>S | No | gnomAD | |
| rs1469918803 | 46 | G>E | No |
TOPMed gnomAD |
|
| rs2055033491 | 47 | E>A | No | TOPMed | |
| rs1464487148 | 47 | E>D | No |
TOPMed gnomAD |
|
| rs770364615 | 49 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs1487768265 | 49 | G>R | No | TOPMed | |
| rs1487768265 | 49 | G>S | No | TOPMed | |
| rs746310894 | 50 | C>W | No |
ExAC gnomAD |
|
| rs1442325529 | 52 | A>V | No |
TOPMed gnomAD |
|
| rs372951506 | 53 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372951506 | 53 | P>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1566774546 | 55 | P>L | No | Ensembl | |
| rs758078648 | 56 | Y>C | No |
TOPMed gnomAD |
|
| rs758078648 | 56 | Y>F | No |
TOPMed gnomAD |
|
| rs1435649576 | 57 | W>* | No | TOPMed | |
| rs1356997860 | 57 | W>R | No | gnomAD | |
| rs1293938741 | 58 | T>A | No | TOPMed | |
| rs778196126 | 58 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs758632490 | 60 | V>M | No |
ExAC gnomAD |
|
| rs778812776 | 61 | F>L | No |
ExAC gnomAD |
|
| rs2055031985 | 62 | E>G | No | gnomAD | |
| rs755066097 | 62 | E>K | No |
ExAC gnomAD |
|
| rs755066097 | 62 | E>Q | No |
ExAC gnomAD |
|
| rs1165857994 | 64 | E>* | No | gnomAD | |
| rs933666193 | 64 | E>G | No | Ensembl | |
| rs1165857994 | 64 | E>Q | No | gnomAD | |
| rs1423200437 | 65 | A>S | No |
TOPMed gnomAD |
|
| rs1423200437 | 65 | A>T | No |
TOPMed gnomAD |
|
| rs1224436134 | 66 | A>P | No |
TOPMed gnomAD |
|
| rs2139877130 | 66 | A>V | No | Ensembl | |
| rs1384198599 | 67 | G>S | No | gnomAD | |
| rs766300372 | 69 | D>H | No |
ExAC gnomAD |
|
| rs766300372 | 69 | D>N | No |
ExAC gnomAD |
|
| rs1239816073 | 70 | E>G | No | gnomAD | |
| rs1450980919 | 70 | E>K | No |
TOPMed gnomAD |
|
| rs867624749 | 72 | T>N | No | Ensembl | |
| rs1340832360 | 72 | T>P | No | gnomAD | |
| TCGA novel | 73 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750161590 | 73 | L>P | No |
ExAC gnomAD |
|
| rs750161590 | 73 | L>Q | No |
ExAC gnomAD |
|
| rs2055030937 | 74 | R>L | No | TOPMed | |
| rs1039356391 | 76 | G>D | No | Ensembl | |
| rs2055030657 | 77 | D>N | No | gnomAD | |
| rs2055030099 | 79 | V>M | No | Ensembl | |
|
COSM1370861 rs2055029934 |
83 | S>F | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated gnomAD |
| rs2055029934 | 83 | S>Y | No | gnomAD | |
| rs1178484450 | 84 | K>E | No | TOPMed | |
| rs1178484450 | 84 | K>Q | No | TOPMed | |
| rs763560050 | 84 | K>R | No |
ExAC gnomAD |
|
| rs377295870 | 85 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs777340994 | 86 | S>L | No |
ExAC gnomAD |
|
| rs1365301547 | 87 | Q>* | No | gnomAD | |
| rs984396831 | 87 | Q>H | No | Ensembl | |
| rs374040206 | 89 | S>A | No |
ESP TOPMed |
|
| rs771546928 | 90 | G>D | No |
ExAC gnomAD |
|
| rs929825886 | 91 | D>N | No | Ensembl | |
| rs1416948546 | 93 | G>A | No | gnomAD | |
| rs2055029180 | 95 | W>C | No |
TOPMed gnomAD |
|
| rs747386773 | 96 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2055029099 | 96 | T>P | No | Ensembl | |
| rs747386773 | 96 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs1594820493 | 97 | G>A | No | Ensembl | |
| rs1270928131 | 97 | G>R | No |
TOPMed gnomAD |
|
| rs2055028671 | 101 | Q>R | No | Ensembl | |
| rs1207052488 | 102 | R>P | No | gnomAD | |
| rs1443174732 | 102 | R>W | No | TOPMed | |
| rs779167690 | 103 | V>L | No |
ExAC gnomAD |
|
| rs779167690 | 103 | V>M | No |
ExAC gnomAD |
|
| rs867165007 | 104 | G>D | No | gnomAD | |
| rs534274868 | 104 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs867165007 | 104 | G>V | No | gnomAD | |
| rs1293145353 | 106 | F>L | No |
TOPMed gnomAD |
|
| rs1662122148 | 107 | P>S | No | TOPMed | |
| rs1423311491 | 110 | Y>C | No | gnomAD | |
| rs567260112 | 110 | Y>H | No |
1000Genomes ExAC gnomAD |
|
| rs2055027804 | 112 | T>I | No | TOPMed | |
| rs896773882 | 112 | T>P | No | Ensembl | |
| rs1352384791 | 113 | P>L | No | gnomAD | |
| rs1414509681 | 114 | R>H | No | gnomAD | |
| rs1414509681 | 114 | R>L | No | gnomAD | |
| rs549400463 | 115 | S>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs549400463 | 115 | S>N | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs531286700 | 115 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs549400463 | 115 | S>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763794847 | 116 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs751209048 | 116 | A>P | No |
ExAC gnomAD |
|
| rs1594820381 | 117 | F>V | No | Ensembl | |
| rs775078243 | 118 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs1479564522 | 118 | S>P | No | gnomAD | |
| rs1204047440 | 119 | S>G | No | gnomAD | |
| rs1594820347 | 119 | S>T | No | gnomAD | |
| rs1443240521 | 120 | R>H | No | gnomAD | |
| rs2055026817 | 122 | Q>E | No | TOPMed | |
| rs1265024823 | 122 | Q>H | No | gnomAD | |
| rs766994142 | 123 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1323442210 | 123 | P>L | No | gnomAD | |
| rs766994142 | 123 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs932015451 | 124 | G>S | No |
TOPMed gnomAD |
|
| rs2055026498 | 125 | G>S | No | Ensembl | |
| rs1297994886 | 126 | E>G | No | gnomAD | |
| rs1371678929 | 126 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1410806280 | 128 | P>S | No |
TOPMed gnomAD |
|
| rs1410806280 | 128 | P>T | No |
TOPMed gnomAD |
|
| rs773837469 | 130 | C>F | No |
ExAC TOPMed gnomAD |
|
| rs2055026195 | 130 | C>W | No | TOPMed | |
| rs772365254 | 132 | P>Q | No |
ExAC gnomAD |
|
| rs1334040534 | 132 | P>S | No | gnomAD | |
| rs1334040534 | 132 | P>T | No | gnomAD | |
| rs2055025903 | 134 | I>T | No |
TOPMed gnomAD |
|
| rs748544153 | 134 | I>V | No |
ExAC gnomAD |
|
| rs1161255136 | 135 | Q>R | No | gnomAD | |
| rs201937853 | 137 | L>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs961518449 | 142 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
|
COSM1658963 rs775097885 |
142 | A>V | salivary_gland [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2054924771 | 143 | E>A | No |
TOPMed gnomAD |
|
| rs774711179 | 143 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs2054924638 | 145 | T>I | No |
TOPMed gnomAD |
|
| rs2054924471 | 149 | I>V | No | Ensembl | |
| rs2054924371 | 150 | I>V | No | Ensembl | |
| rs2139861144 | 151 | G>D | No | Ensembl | |
| rs1162717009 | 153 | G>A | No | gnomAD | |
| rs2139861117 | 154 | G>S | No | Ensembl | |
| rs2054924171 | 155 | F>Y | No | TOPMed | |
| rs1362461090 | 156 | G>E | No |
TOPMed gnomAD |
|
| COSM3690174 | 157 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1566769765 | 158 | V>F | No | Ensembl | |
| TCGA novel | 158 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781046152 | 159 | Y>C | No |
ExAC TOPMed gnomAD |
|
|
COSM3497602 rs770841109 |
160 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs142171628 COSM240595 |
160 | R>H | prostate [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs1198599847 | 162 | F>L | No | TOPMed | |
| rs758110290 | 164 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs777581351 | 164 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs752441948 | 165 | G>R | No |
ExAC TOPMed gnomAD |
|
|
rs1184494856 COSM433317 |
166 | D>N | Variant assessed as Somatic; MODERATE impact. breast [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs753342747 | 168 | V>F | No |
ExAC gnomAD |
|
| rs753342747 | 168 | V>I | No |
ExAC gnomAD |
|
| rs762342117 | 173 | A>S | No |
ExAC gnomAD |
|
| rs2054923471 | 174 | R>C | No |
TOPMed gnomAD |
|
| rs764569241 | 174 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs764569241 | 174 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs200295742 | 175 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763271384 | 175 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs1343579550 | 175 | H>Y | No |
TOPMed gnomAD |
|
| rs934732804 | 176 | D>H | No |
TOPMed gnomAD |
|
| rs934732804 | 176 | D>N | No |
TOPMed gnomAD |
|
| rs1460687517 | 178 | D>V | No | gnomAD | |
| rs1457913712 | 179 | E>K | No | gnomAD | |
| rs1171296299 | 180 | D>A | No |
TOPMed gnomAD |
|
| rs776702942 | 180 | D>E | No |
ExAC gnomAD |
|
| rs374413255 | 180 | D>N | No |
ESP ExAC gnomAD |
|
| rs374413255 | 180 | D>Y | No |
ESP ExAC gnomAD |
|
| rs183066238 | 181 | I>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs868151792 | 181 | I>V | No |
TOPMed gnomAD |
|
| rs1433809496 | 185 | I>M | No | Ensembl | |
| rs2054922225 | 185 | I>T | No |
TOPMed gnomAD |
|
| rs1199135635 | 185 | I>V | No |
TOPMed gnomAD |
|
| rs1438809940 | 186 | E>G | No | gnomAD | |
| rs1252185407 | 187 | N>S | No | gnomAD | |
| rs923437687 | 188 | V>I | No |
TOPMed gnomAD |
|
| rs747019799 | 189 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs777544362 | 189 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2054921995 | 190 | Q>R | No | Ensembl | |
| rs376594979 | 196 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs376594979 | 196 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1377097688 | 197 | M>T | No | gnomAD | |
| rs1239475856 | 199 | K>N | No | gnomAD | |
| rs2054921684 | 199 | K>R | No | Ensembl | |
| rs1337269416 | 200 | H>D | No | gnomAD | |
| rs1337269416 | 200 | H>Y | No | gnomAD | |
| rs754612694 | 201 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs1817600785 | 201 | P>L | No | gnomAD | |
| rs1817600785 | 201 | P>R | No | gnomAD | |
| rs1389834144 | 202 | N>S | No | gnomAD | |
| rs748780174 | 203 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2054921353 | 204 | I>T | No | Ensembl | |
| rs2054921386 | 204 | I>V | No |
TOPMed gnomAD |
|
| rs1471031732 | 205 | A>T | No | gnomAD | |
| rs1481945375 | 206 | L>V | No | Ensembl | |
| rs755556091 | 209 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs755556091 | 209 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs752148458 | 210 | C>R | No |
ExAC TOPMed gnomAD |
|
| rs2054921073 | 211 | L>M | No |
TOPMed gnomAD |
|
| rs774200566 | 211 | L>P | No | Ensembl | |
| rs2054920977 | 212 | K>T | No | gnomAD | |
| rs1447846323 | 213 | E>D | No | gnomAD | |
| rs937786253 | 214 | P>A | No | gnomAD | |
| rs764502562 | 215 | N>S | No | ExAC | |
| rs1365856572 | 216 | L>F | No | gnomAD | |
| rs2139860601 | 218 | L>M | No | Ensembl | |
| rs2054920637 | 219 | V>G | No | TOPMed | |
| rs1180122484 | 219 | V>I | No | gnomAD | |
| rs376560990 | 220 | M>I | No | Ensembl | |
| rs1418078621 | 220 | M>L | No | gnomAD | |
| rs1418078621 | 220 | M>V | No | gnomAD | |
| rs2054920478 | 221 | E>* | No | TOPMed | |
| rs2054920429 | 222 | F>S | No | Ensembl | |
| rs199845016 | 223 | A>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1323406 | 223 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs148675299 | 224 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA gnomAD |
| rs373287003 | 224 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373287003 | 224 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373287003 | 224 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs776593654 | 225 | G>* | No |
ExAC gnomAD |
|
| rs766524571 | 226 | G>E | No |
ExAC gnomAD |
|
| rs1402721137 | 228 | L>W | No |
TOPMed gnomAD |
|
| rs2054920048 | 229 | N>H | No | TOPMed | |
| rs2054920013 | 229 | N>S | No | TOPMed | |
| rs1352768289 | 231 | V>M | No | gnomAD | |
| rs144036386 | 233 | S>P | No |
ESP ExAC gnomAD |
|
| rs1316058516 | 234 | G>R | No | gnomAD | |
| rs747590864 | 235 | K>Q | No |
ExAC gnomAD |
|
| rs774250345 | 236 | R>S | No |
ExAC gnomAD |
|
| rs768445956 | 237 | I>V | No |
ExAC gnomAD |
|
| rs961113927 | 238 | P>H | No | Ensembl | |
|
COSM1678109 rs373007964 |
238 | P>T | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| COSM3497599 | 239 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs139396475 | 240 | D>H | No |
ESP ExAC gnomAD |
|
| rs2054919263 | 241 | I>V | No | Ensembl | |
| rs1442420564 | 242 | L>V | No | gnomAD | |
| rs1241922530 | 244 | N>H | No | gnomAD | |
| rs2139860361 | 244 | N>K | No | Ensembl | |
| rs2054919110 | 245 | W>G | No | Ensembl | |
| rs1331616996 | 245 | W>L | No | TOPMed | |
|
COSM1370860 rs202244940 |
246 | A>T | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| VAR_040698 | 246 | A>V | a metastatic melanoma sample; somatic mutation [UniProt] | No | UniProt |
| rs2054919008 | 247 | V>M | No | TOPMed | |
| rs778262427 | 249 | I>F | No |
ExAC gnomAD |
|
| rs1259641477 | 249 | I>T | No |
TOPMed gnomAD |
|
| TCGA novel | 250 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2054918855 | 253 | M>I | No | Ensembl | |
| rs989501118 | 255 | Y>* | No | TOPMed | |
| rs753087369 | 256 | L>F | No |
ExAC gnomAD |
|
| rs2054918691 | 257 | H>N | No | Ensembl | |
| rs758741981 | 258 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs1261510841 | 258 | D>H | No |
TOPMed gnomAD |
|
| rs758741981 | 258 | D>V | No |
ExAC TOPMed gnomAD |
|
| rs1261510841 | 258 | D>Y | No |
TOPMed gnomAD |
|
|
rs1266675185 COSM459120 |
259 | E>K | cervix Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs1248391787 | 260 | A>S | No | gnomAD | |
| rs1283608694 | 261 | I>T | No | gnomAD | |
| rs753129930 | 261 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs766499534 | 263 | P>A | No |
ExAC gnomAD |
|
| rs868010261 | 263 | P>H | No | TOPMed | |
| rs868010261 | 263 | P>L | No | TOPMed | |
| rs200277034 | 265 | I>V | No |
1000Genomes ESP TOPMed gnomAD |
|
| rs760824974 | 267 | R>C | No |
ExAC gnomAD |
|
| rs773099721 | 267 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs773099721 | 267 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs2054917957 | 268 | D>N | No | TOPMed | |
| rs1167974510 | 272 | S>N | No |
TOPMed gnomAD |
|
| rs2054369636 | 274 | I>M | No | gnomAD | |
| rs770498608 | 277 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1384314056 | 281 | E>V | No | gnomAD | |
| rs2054369331 | 282 | N>H | No | Ensembl | |
| rs557062833 | 285 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2054369061 | 287 | N>S | No | TOPMed | |
| rs2054369013 | 288 | K>R | No | gnomAD | |
| rs2054368917 | 289 | I>F | No | Ensembl | |
| rs2054368857 | 292 | I>V | No | Ensembl | |
| rs2054368810 | 294 | D>V | No | TOPMed | |
| rs1378663115 | 295 | F>C | No |
TOPMed gnomAD |
|
| rs1378663115 | 295 | F>S | No |
TOPMed gnomAD |
|
| rs551815229 | 298 | A>D | No |
1000Genomes ExAC gnomAD |
|
| rs551815229 | 298 | A>V | No |
1000Genomes ExAC gnomAD |
|
| rs947471527 | 299 | R>Q | No |
TOPMed gnomAD |
|
|
COSM3497598 rs757467747 |
299 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1490598202 | 301 | W>* | No |
TOPMed gnomAD |
|
| rs2054368309 | 302 | H>Q | No | gnomAD | |
|
COSM195176 rs2054368374 |
302 | H>Y | large_intestine [Cosmic] | No |
cosmic curated Ensembl |
| rs914738850 | 303 | R>* | No | gnomAD | |
| rs988982557 | 303 | R>Q | No |
TOPMed gnomAD |
|
| rs2054368140 | 304 | T>I | No | TOPMed | |
| rs764096308 | 306 | K>T | No |
ExAC gnomAD |
|
| rs1249020055 | 307 | M>I | No | TOPMed | |
| rs762755525 | 308 | S>G | No |
ExAC gnomAD |
|
| rs2054367932 | 308 | S>T | No | Ensembl | |
| rs752555260 | 309 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs752555260 | 309 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs370660341 | 309 | A>V | No |
1000Genomes ExAC gnomAD |
|
| TCGA novel | 311 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776326945 | 311 | G>R | No |
ExAC gnomAD |
|
|
rs1163109119 COSM2136315 |
312 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs1594785849 | 313 | Y>D | No | Ensembl | |
|
COSM698798 rs200962207 |
317 | A>T | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
|
COSM1707529 rs866106057 |
319 | E>K | Variant assessed as Somatic; MODERATE impact. skin [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
|
rs770124979 COSM1370859 |
322 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs776036323 COSM3497597 |
322 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs781577340 | 323 | A>D | No |
ExAC gnomAD |
|
| rs781577340 | 323 | A>V | No |
ExAC gnomAD |
|
| rs757487803 | 324 | S>F | No |
ExAC gnomAD |
|
| COSM6076161 | 325 | M>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs777660953 | 325 | M>T | No |
ExAC TOPMed gnomAD |
|
| rs747112872 | 325 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs2054366788 | 326 | F>S | No | gnomAD | |
| rs758479516 | 329 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1054281922 | 330 | S>N | No | TOPMed | |
| rs752614049 | 331 | D>N | No |
ExAC gnomAD |
|
| rs1391933994 | 332 | V>M | No |
TOPMed gnomAD |
|
| rs1566740567 | 335 | Y>C | No | Ensembl | |
| rs754875078 | 337 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs2054203715 | 338 | L>P | No | Ensembl | |
| rs1300358993 | 342 | L>V | No | gnomAD | |
| rs766212301 | 343 | L>V | No |
ExAC gnomAD |
|
| rs755873556 | 347 | V>M | No |
ExAC gnomAD |
|
|
COSM172300 rs1299953155 |
350 | R>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
cosmic curated NCI-TCGA gnomAD |
| rs1464177559 | 350 | R>Q | No | gnomAD | |
| rs1177731102 | 352 | I>T | No | gnomAD | |
| rs1376222649 | 353 | D>G | No | gnomAD | |
|
COSM3497596 rs2054203012 |
354 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1419294627 | 356 | A>P | No | gnomAD | |
| rs751015012 | 358 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs1263257672 | 359 | Y>S | No | gnomAD | |
| rs1255130138 | 363 | M>K | No |
TOPMed gnomAD |
|
| rs1255130138 | 363 | M>R | No |
TOPMed gnomAD |
|
| TCGA novel | 363 | M>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1255130138 | 363 | M>T | No |
TOPMed gnomAD |
|
| rs141503439 | 363 | M>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs760067989 | 364 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs2054202465 | 366 | L>F | No | Ensembl | |
|
rs879195926 COSM4757188 |
367 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl NCI-TCGA |
|
COSM1678107 rs879195926 |
367 | A>T | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| rs2054202227 | 369 | P>S | No | TOPMed | |
| rs2054202113 | 370 | I>T | No | TOPMed | |
| rs771460603 | 370 | I>V | No |
ExAC gnomAD |
|
| COSM4901519 | 371 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs983821213 | 372 | S>C | No | TOPMed | |
| rs761093575 | 373 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs1017260225 | 373 | T>M | No |
TOPMed gnomAD |
|
| rs1022747447 | 375 | P>S | No |
TOPMed gnomAD |
|
| rs748211903 | 377 | P>R | No | ExAC | |
| rs2054201696 | 378 | F>C | No | Ensembl | |
| rs779005476 | 378 | F>L | No |
ExAC gnomAD |
|
| rs1455164008 | 378 | F>L | No |
TOPMed gnomAD |
|
| rs768701260 | 379 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs749264998 | 379 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1454053508 | 380 | K>R | No |
TOPMed gnomAD |
|
| rs779963503 | 382 | M>V | No |
ExAC gnomAD |
|
| rs755963592 | 383 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1387661138 | 384 | D>N | No | TOPMed | |
| rs1230073684 | 385 | C>Y | No | gnomAD | |
| rs372283795 | 389 | D>N | No | gnomAD | |
| rs745679893 | 390 | P>L | No |
ExAC gnomAD |
|
| TCGA novel | 390 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs756976430 | 392 | S>* | No |
ExAC gnomAD |
|
| rs756976430 | 392 | S>L | No |
ExAC gnomAD |
|
| rs780991502 | 392 | S>P | No |
ExAC gnomAD |
|
| rs2054188620 | 393 | R>* | No | TOPMed | |
| rs2054188575 | 393 | R>Q | No | TOPMed | |
| rs1399847365 | 397 | T>A | No |
TOPMed gnomAD |
|
| rs776638628 | 397 | T>M | No |
ExAC gnomAD |
|
| TCGA novel | 399 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758038259 | 400 | L>V | No |
ExAC gnomAD |
|
| rs1445416439 | 403 | L>V | No | TOPMed | |
| rs1389314441 | 406 | I>R | No |
TOPMed gnomAD |
|
|
rs1389314441 COSM4927513 |
406 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1355173741 | 409 | S>A | No |
TOPMed gnomAD |
|
| rs202234944 | 410 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs756598509 | 410 | G>S | No |
ExAC gnomAD |
|
| rs1594776742 | 411 | F>L | No | TOPMed | |
| rs1295582220 | 412 | F>Y | No | TOPMed | |
| COSM3497595 | 413 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4851482 | 413 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1480665308 | 414 | M>I | No |
TOPMed gnomAD |
|
| rs1223995350 | 417 | D>A | No |
TOPMed gnomAD |
|
| rs1223995350 | 417 | D>G | No |
TOPMed gnomAD |
|
| rs2054187492 | 420 | H>Y | No | TOPMed | |
| rs764245442 | 423 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs764245442 | 423 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2054187206 | 424 | D>N | No | Ensembl | |
| rs2054187150 | 425 | N>D | No | Ensembl | |
| rs1566739645 | 425 | N>S | No |
TOPMed gnomAD |
|
| rs1485836848 | 426 | W>G | No |
TOPMed gnomAD |
|
| rs865839912 | 426 | W>L | No | Ensembl | |
| rs1204907636 | 427 | K>Q | No | Ensembl | |
| rs201699297 | 428 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs763022930 | 428 | H>Y | No |
ExAC gnomAD |
|
| TCGA novel | 429 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs868595607 | 429 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1218597263 | 431 | Q>R | No | gnomAD | |
| rs1240268098 | 432 | E>K | No | Ensembl | |
| rs1475079727 | 433 | M>I | No |
TOPMed gnomAD |
|
| COSM698799 | 435 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs551542312 | 436 | Q>E | No |
1000Genomes ExAC gnomAD |
|
| rs2054186502 | 437 | L>R | No | Ensembl | |
| rs745866425 | 438 | R>G | No |
ExAC gnomAD |
|
| rs1167954560 | 439 | A>V | No |
TOPMed gnomAD |
|
| rs1483855404 | 444 | L>F | No | gnomAD | |
| rs1483855404 | 444 | L>I | No | gnomAD | |
| rs201808611 | 445 | R>C | No |
ExAC TOPMed gnomAD |
|
|
COSM957428 rs150546098 |
445 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs150546098 | 445 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 446 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs754574540 | 446 | T>I | No |
ExAC TOPMed gnomAD |
|
| COSM3497594 | 447 | W>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2054088049 | 447 | W>C | No | Ensembl | |
| rs748690112 | 447 | W>S | No |
ExAC gnomAD |
|
| rs781725690 | 448 | E>K | No |
ExAC gnomAD |
|
| rs1351065757 | 449 | E>K | No | gnomAD | |
| rs1301058346 | 452 | T>M | No |
TOPMed gnomAD |
|
| rs148357774 | 453 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs752979159 COSM3497593 |
453 | R>W | Variant assessed as Somatic; MODERATE impact. pancreas [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs376843564 | 455 | A>P | No | ESP | |
| rs2054087257 | 455 | A>V | No |
TOPMed gnomAD |
|
| rs1594771709 | 456 | L>P | No | Ensembl | |
|
COSM1707528 rs549151946 |
456 | L>V | skin [Cosmic] | No |
cosmic curated 1000Genomes ExAC TOPMed gnomAD |
| rs983867368 | 458 | Q>R | No |
TOPMed gnomAD |
|
| COSM4052111 | 459 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1273922532 | 460 | N>K | No | TOPMed | |
| rs931040989 | 461 | Q>E | No | TOPMed | |
| rs1416087523 | 463 | E>K | No | gnomAD | |
| rs569837216 | 464 | L>R | No |
1000Genomes ExAC gnomAD |
|
| rs1483217886 | 466 | R>G | No |
TOPMed gnomAD |
|
| rs1220234795 | 466 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1483217886 | 466 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
VAR_040699 COSM20543 rs773256562 |
467 | R>C | large_intestine stomach a gastric adenocarcinoma sample; somatic mutation [Cosmic, UniProt] | No |
cosmic curated UniProt ExAC dbSNP gnomAD |
| rs373021591 | 467 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs774068235 | 468 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs761687543 | 468 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs768128204 | 469 | E>G | No |
ExAC gnomAD |
|
| rs1227606511 | 470 | Q>R | No | gnomAD | |
| rs143332721 | 471 | E>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs779381219 | 471 | E>K | No |
ExAC gnomAD |
|
| rs145226088 | 474 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs752943272 | 475 | R>L | No |
ExAC gnomAD |
|
| rs752943272 | 475 | R>Q | No |
ExAC gnomAD |
|
| rs1297774580 | 475 | R>W | No |
TOPMed gnomAD |
|
| rs1351635124 | 476 | E>Q | No | gnomAD | |
| COSM6076162 | 477 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1423743195 | 477 | I>T | No | gnomAD | |
| rs779198242 | 477 | I>V | No |
ExAC gnomAD |
|
| rs755109645 | 479 | I>V | No |
ExAC gnomAD |
|
| rs1477069784 | 481 | E>G | No | gnomAD | |
| rs959910708 | 481 | E>K | No |
TOPMed gnomAD |
|
| rs754071935 | 482 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs754071935 | 482 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs1262960434 COSM1370857 |
482 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine central_nervous_system [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs766411031 | 485 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs375161594 | 486 | I>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs767507468 | 487 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs767507468 | 487 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs1318479351 | 488 | I>V | No |
TOPMed gnomAD |
|
| rs1035216817 | 489 | H>Q | No | Ensembl | |
| rs761708031 | 489 | H>Y | No |
ExAC gnomAD |
|
| rs2054084481 | 490 | Q>E | No | Ensembl | |
| rs1216752547 | 492 | C>S | No | gnomAD | |
| TCGA novel | 494 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs370542202 | 494 | E>K | No |
ESP ExAC gnomAD |
|
| rs1283704730 | 495 | K>N | No | gnomAD | |
| rs1387502876 | 496 | P>A | No |
TOPMed gnomAD |
|
| rs2054084063 | 496 | P>L | No | gnomAD | |
| rs1387502876 | 496 | P>S | No |
TOPMed gnomAD |
|
| rs1387502876 | 496 | P>T | No |
TOPMed gnomAD |
|
|
VAR_040700 rs56196343 |
497 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
UniProt ExAC NCI-TCGA TOPMed dbSNP gnomAD |
| rs1291849494 | 497 | R>W | No | gnomAD | |
| rs2054083783 | 500 | K>T | No | Ensembl | |
|
COSM4643859 rs547034010 |
501 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs769357313 | 501 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs769357313 | 501 | R>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 502 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs111385383 | 503 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1423274051 | 504 | K>N | No | gnomAD | |
| rs1226734356 | 504 | K>R | No | Ensembl | |
| rs772500475 | 505 | F>Y | No |
ExAC gnomAD |
|
| rs2139732815 | 507 | K>N | No | Ensembl | |
| rs2054083024 | 509 | R>Q | No | TOPMed | |
| rs779197132 | 509 | R>W | No |
ExAC TOPMed gnomAD |
|
| COSM3497590 | 511 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2054082978 | 512 | L>P | No | TOPMed | |
| rs1566735468 | 513 | K>M | No | TOPMed | |
| rs373570314 | 513 | K>N | No |
ESP ExAC gnomAD |
|
| rs1566735468 | 513 | K>R | No | TOPMed | |
| rs1340570500 | 515 | G>C | No |
TOPMed gnomAD |
|
| rs1340570500 | 515 | G>S | No |
TOPMed gnomAD |
|
| rs1452334418 | 516 | N>I | No | gnomAD | |
| rs1452334418 | 516 | N>S | No | gnomAD | |
| rs147037059 | 517 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs200816838 | 517 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4052110 | 519 | S>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2054082411 | 520 | L>P | No | gnomAD | |
|
rs2139732651 COSM698800 |
521 | P>S | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs780432308 | 523 | D>E | No |
ExAC gnomAD |
|
| COSM6076163 | 523 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1238120254 | 529 | T>M | No |
TOPMed gnomAD |
|
| TCGA novel | 530 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2054050071 | 532 | A>G | No | Ensembl | |
| TCGA novel | 533 | S>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs2054049961 COSM3497589 |
533 | S>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs141502233 | 534 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs141502233 | 534 | P>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs147874450 | 536 | M>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs745969786 | 536 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1223743642 | 537 | D>E | No | TOPMed | |
| rs2054049593 | 537 | D>G | No | Ensembl | |
| rs1038858386 | 537 | D>H | No |
TOPMed gnomAD |
|
| rs2054049263 | 540 | K>N | No |
TOPMed gnomAD |
|
| rs1312655852 | 540 | K>Q | No | gnomAD | |
| rs751515326 | 542 | L>R | No |
ExAC gnomAD |
|
| rs1230648389 | 543 | I>T | No | TOPMed | |
| rs2054048997 | 544 | N>H | No | TOPMed | |
| rs140979438 | 544 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs74907260 | 544 | N>T | No |
ExAC gnomAD |
|
| COSM4920552 | 545 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs758192028 | 546 | R>C | No |
TOPMed gnomAD |
|
| rs564950962 | 546 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs564950962 | 546 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM957426 | 549 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776237182 | 550 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs759137263 | 550 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs759137263 | 550 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs545676302 | 551 | A>G | No |
1000Genomes ExAC gnomAD |
|
| rs369129444 | 552 | S>I | No |
ESP ExAC gnomAD |
|
| rs1359037479 | 553 | P>A | No |
TOPMed gnomAD |
|
| rs1349978604 | 554 | T>A | No | gnomAD | |
| rs2054048052 | 554 | T>N | No | TOPMed | |
| rs1293393097 | 555 | I>V | No | gnomAD | |
| rs769145256 | 556 | I>F | No |
ExAC TOPMed gnomAD |
|
|
COSM3497588 rs2054047843 |
557 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs749649763 | 558 | R>C | No |
ExAC gnomAD |
|
| rs775951489 | 558 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1594769632 | 560 | R>* | No | TOPMed | |
| rs770263201 | 561 | A>T | No |
ExAC gnomAD |
|
| rs746130985 | 561 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs781556899 | 562 | I>V | No |
ExAC gnomAD |
|
| rs1241745614 | 565 | T>R | No | gnomAD | |
| rs1278842635 | 566 | P>R | No | gnomAD | |
| rs2054016087 | 566 | P>S | No | Ensembl | |
| rs2054015988 | 569 | S>T | No | gnomAD | |
| rs1465795351 | 570 | S>G | No | Ensembl | |
| rs779950314 | 572 | T>I | No |
ExAC gnomAD |
|
| rs2054015773 | 573 | W>R | No | Ensembl | |
| rs755837546 | 574 | G>S | No |
ExAC gnomAD |
|
| rs745598440 | 576 | S>R | No |
ExAC gnomAD |
|
| rs756837051 | 577 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs1566732539 | 577 | S>P | No | Ensembl | |
| rs372484455 | 579 | V>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs372484455 | 579 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150993424 | 580 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs150993424 | 580 | P>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2054015171 | 580 | P>S | No |
TOPMed gnomAD |
|
| rs2139720789 | 582 | E>D | No | Ensembl | |
| rs1278796124 | 582 | E>K | No | gnomAD | |
| rs1209249796 | 584 | G>E | No |
TOPMed gnomAD |
|
| rs1436945562 | 584 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1463462503 | 585 | E>A | No | TOPMed | |
| rs1463462503 | 585 | E>G | No | TOPMed | |
| rs1485126606 | 585 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1463462503 | 585 | E>V | No | TOPMed | |
| rs2139720707 | 586 | E>K | No | Ensembl | |
| COSM6140978 | 588 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs924647813 | 588 | E>Q | No |
TOPMed gnomAD |
|
| rs1350802161 | 588 | E>V | No | gnomAD | |
| COSM3497586 | 591 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs760936170 | 592 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs760936170 | 592 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs372081680 | 593 | K>E | No |
ESP TOPMed gnomAD |
|
| rs1423978157 | 594 | K>Q | No |
TOPMed gnomAD |
|
| rs2054014194 | 595 | K>E | No | gnomAD | |
| rs199721479 | 595 | K>R | No | 1000Genomes | |
| rs142599599 | 597 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs139436577 | 597 | R>Q | No |
ESP TOPMed |
|
| rs142599599 | 597 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs572761925 COSM1678105 |
598 | T>M | Variant assessed as Somatic; MODERATE impact. central_nervous_system haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs865971816 | 600 | G>R | No | Ensembl | |
| rs1160498676 | 601 | P>L | No |
TOPMed gnomAD |
|
| rs745589833 | 603 | T>M | No |
ExAC gnomAD |
|
| rs780897561 | 604 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs756926877 | 605 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs374508438 | 606 | Q>E | No |
ESP gnomAD |
|
| rs746538544 | 608 | E>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 608 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs777473872 | 609 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs2054013208 | 609 | L>P | No | TOPMed | |
| rs2054013208 | 609 | L>R | No | TOPMed | |
| rs754433823 | 611 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs2054012869 | 613 | D>G | No |
TOPMed gnomAD |
|
| rs2054012869 | 613 | D>V | No |
TOPMed gnomAD |
|
|
rs1479227792 COSM48535 |
615 | G>A | lung [Cosmic] | No |
cosmic curated gnomAD |
| rs370186142 | 615 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1484678828 | 616 | S>C | No | TOPMed | |
| COSM3497585 | 616 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1305952794 | 617 | P>S | No | gnomAD | |
| TCGA novel | 618 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053981746 | 618 | Q>R | No | Ensembl | |
| TCGA novel | 618 | Q>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750923784 | 619 | R>K | No |
ExAC gnomAD |
|
| rs781580404 | 620 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs757616738 | 620 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs757616738 | 620 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs1196340028 | 621 | E>G | No | Ensembl | |
| TCGA novel | 622 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053981425 | 623 | A>T | No | Ensembl | |
| rs1323979524 | 623 | A>V | No | gnomAD | |
| rs1427780034 | 624 | N>D | No |
TOPMed gnomAD |
|
| rs2053981188 | 624 | N>I | No | TOPMed | |
| rs1427780034 | 624 | N>Y | No |
TOPMed gnomAD |
|
| rs1401740868 | 629 | P>A | No | gnomAD | |
| rs764239615 | 629 | P>L | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 629 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1460828187 | 630 | S>L | No | gnomAD | |
| rs943230880 | 634 | H>R | No | TOPMed | |
| rs1176258852 | 635 | F>L | No |
TOPMed gnomAD |
|
| rs752675446 | 636 | H>Y | No |
ExAC gnomAD |
|
| rs765238955 | 638 | G>D | No |
ExAC gnomAD |
|
| rs1392714580 | 638 | G>S | No | gnomAD | |
| rs765238955 | 638 | G>V | No |
ExAC gnomAD |
|
| rs143859920 | 639 | L>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs1260848025 | 639 | L>P | No | TOPMed | |
| rs2053957418 | 640 | K>R | No | TOPMed | |
| rs752941521 | 641 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs752941521 | 641 | S>Y | No |
ExAC TOPMed gnomAD |
|
| rs759655188 | 643 | V>A | No |
ExAC gnomAD |
|
| rs759655188 | 643 | V>E | No |
ExAC gnomAD |
|
| rs1426424963 | 644 | D>N | No |
TOPMed gnomAD |
|
|
VAR_040701 rs34322726 |
646 | Y>C | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs760579283 | 647 | K>E | No |
ExAC gnomAD |
|
| TCGA novel | 647 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1313854302 | 649 | W>R | No | gnomAD | |
| rs758241616 | 650 | S>L | No |
ExAC TOPMed gnomAD |
|
| rs2053956552 | 651 | S>Y | No | TOPMed | |
| rs774052286 | 654 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs188387961 | 655 | N>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs188387961 | 655 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1452952140 | 657 | V>G | No | gnomAD | |
| rs370461429 | 657 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1299305917 | 658 | K>R | No | TOPMed | |
| rs1369039993 | 659 | G>C | No | gnomAD | |
| TCGA novel | 660 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143943391 | 661 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 662 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053955915 | 662 | S>N | No | Ensembl | |
| rs758757927 | 662 | S>R | No | ExAC | |
| rs116289537 | 664 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs116289537 | 664 | P>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1221091703 | 666 | L>V | No |
TOPMed gnomAD |
|
| rs2053955539 | 667 | P>L | No | Ensembl | |
| rs575813257 | 668 | G>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs575813257 | 668 | G>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1263923343 | 668 | G>R | No | gnomAD | |
| rs575813257 | 668 | G>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1263923343 | 668 | G>W | No | gnomAD | |
| rs1202780176 | 671 | S>G | No | gnomAD | |
| rs200983878 | 671 | S>N | No | gnomAD | |
| rs200983878 | 671 | S>T | No | gnomAD | |
| rs1265207599 | 672 | L>P | No |
TOPMed gnomAD |
|
| rs767285366 | 673 | M>V | No |
ExAC gnomAD |
|
| rs2053954988 | 675 | M>T | No | gnomAD | |
| rs2053955030 | 675 | M>V | No | Ensembl | |
| rs2053940309 | 676 | E>G | No | Ensembl | |
| rs764793311 | 677 | D>H | No |
ExAC TOPMed gnomAD |
|
| rs764793311 | 677 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1281968777 | 678 | E>* | No |
TOPMed gnomAD |
|
| rs1281968777 | 678 | E>K | No |
TOPMed gnomAD |
|
| rs1208307352 | 681 | E>K | No | gnomAD | |
| rs2139706452 | 682 | G>R | No | Ensembl | |
| rs759030999 | 683 | P>Q | No | ExAC | |
| rs1594763628 | 683 | P>S | No | gnomAD | |
| rs772563409 | 684 | G>R | No |
ExAC gnomAD |
|
| rs745505569 | 687 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs185207367 | 689 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM957424 rs147124893 |
689 | R>H | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1594763581 | 692 | H>L | No | Ensembl | |
|
rs1016751326 COSM3886305 |
693 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1433111208 | 693 | S>P | No | gnomAD | |
| rs1236615958 | 694 | P>H | No |
TOPMed gnomAD |
|
| rs1159993774 | 694 | P>S | No |
TOPMed gnomAD |
|
| rs2053938894 | 695 | S>I | No | Ensembl | |
| rs2053938838 | 696 | Q>H | No | gnomAD | |
| rs1475710727 | 702 | P>R | No | gnomAD | |
| rs757166012 | 702 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs757166012 | 702 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs751427569 | 704 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs374128261 | 704 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs931525178 | 705 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs146243565 | 705 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs146243565 | 705 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2139706103 | 706 | G>E | No | Ensembl | |
|
rs2139706115 COSM3497583 |
706 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs1037286156 | 709 | G>D | No |
TOPMed gnomAD |
|
| rs1037286156 | 709 | G>V | No |
TOPMed gnomAD |
|
| rs764811729 | 710 | D>E | No |
ExAC TOPMed gnomAD |
|
|
rs752402687 COSM1300792 |
710 | D>N | urinary_tract [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs765783857 | 713 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs2053937808 | 713 | S>P | No | TOPMed | |
| rs1355113648 | 717 | I>N | No |
TOPMed gnomAD |
|
| rs1248282828 | 718 | H>R | No | gnomAD | |
| rs2053937514 | 719 | E>A | No | Ensembl | |
| rs2139705968 | 719 | E>D | No | Ensembl | |
| rs762423971 | 719 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1339144516 | 720 | E>G | No |
TOPMed gnomAD |
|
| TCGA novel | 720 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053937286 | 721 | P>H | No | gnomAD | |
| rs1594763435 | 721 | P>S | No |
TOPMed gnomAD |
|
| rs373164855 | 722 | T>I | No |
ESP ExAC gnomAD |
|
| rs373164855 | 722 | T>N | No |
ESP ExAC gnomAD |
|
| rs1318180197 | 723 | P>A | No | gnomAD | |
| rs566965450 | 724 | V>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs770105473 | 725 | N>H | No |
ExAC gnomAD |
|
| rs745952959 | 726 | S>L | No |
ExAC gnomAD |
|
| rs143232244 | 728 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs911342222 | 729 | S>I | No |
TOPMed gnomAD |
|
| rs911342222 | 729 | S>N | No |
TOPMed gnomAD |
|
| rs1469824084 | 730 | T>N | No | gnomAD | |
| rs1594763354 | 730 | T>P | No | Ensembl | |
| rs1283773667 | 731 | P>A | No | TOPMed | |
| rs2053936286 | 731 | P>L | No | gnomAD | |
| rs2053936286 | 731 | P>R | No | gnomAD | |
| rs747871429 | 734 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs754709328 | 738 | S>I | No |
ExAC TOPMed gnomAD |
|
| rs754709328 | 738 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs1288432646 | 739 | L>F | No |
TOPMed gnomAD |
|
| TCGA novel | 740 | K>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs990507598 | 741 | R>Q | No |
TOPMed gnomAD |
|
| rs145221753 | 741 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs958153782 | 742 | G>S | No |
TOPMed gnomAD |
|
| rs1461215725 | 742 | G>V | No | gnomAD | |
| rs1172015239 | 743 | G>S | No |
TOPMed gnomAD |
|
| rs1359181904 | 744 | A>G | No |
TOPMed gnomAD |
|
| rs200664597 | 745 | H>P | No |
ExAC gnomAD |
|
| rs755568141 | 746 | H>P | No |
ExAC gnomAD |
|
| rs1193788238 | 747 | R>C | No |
TOPMed gnomAD |
|
|
COSM99078 rs372217059 |
747 | R>H | stomach [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs149935990 | 748 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs375119611 | 748 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs375119611 | 748 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs375119611 | 748 | R>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs759840472 | 750 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1236208108 | 751 | V>M | No | TOPMed | |
| COSM957423 | 753 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs999475384 | 755 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1594763182 | 756 | C>W | No | Ensembl | |
| rs776710305 | 757 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs2053934474 | 757 | G>V | No | TOPMed | |
| rs773137594 | 758 | A>T | No |
ExAC gnomAD |
|
| rs3814874 | 759 | V>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs3814874 | 759 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2053934199 | 760 | L>V | No | TOPMed | |
| rs1253213832 | 761 | A>P | No | TOPMed | |
| rs1253213832 | 761 | A>T | No | TOPMed | |
| rs748056144 | 763 | T>I | No |
ExAC gnomAD |
|
| rs1176403355 | 768 | D>G | No | TOPMed | |
| rs749917098 | 771 | E>* | No |
ExAC gnomAD |
|
| rs764724247 | 771 | E>D | No |
ExAC gnomAD |
|
| rs753220614 | 774 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs1014191193 | 775 | C>Y | No |
TOPMed gnomAD |
|
| rs778571437 | 777 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs778571437 | 777 | L>Q | No |
ExAC TOPMed gnomAD |
|
| rs776922633 | 779 | P>R | No |
ExAC TOPMed gnomAD |
|
| COSM3886304 | 779 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2053932761 | 781 | E>K | No | Ensembl | |
| rs766443716 | 782 | E>K | No |
ExAC gnomAD |
|
| rs201326431 | 783 | P>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs201326431 | 783 | P>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs772109137 | 786 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs772109137 | 786 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs748151841 | 787 | A>P | No |
ExAC gnomAD |
|
| rs748151841 | 787 | A>S | No |
ExAC gnomAD |
|
| rs367998651 | 787 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs768511767 | 788 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs368928316 | 788 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs768511767 | 788 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs779790828 | 789 | E>K | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 792 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769286914 | 794 | R>G | No |
ExAC TOPMed gnomAD |
|
|
COSM195156 rs745488869 |
794 | R>Q | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
|
COSM1196072 rs769286914 |
794 | R>W | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1156877131 | 795 | E>G | No | gnomAD | |
| rs1418240559 | 796 | G>V | No | gnomAD | |
| rs940301306 | 797 | L>P | No |
TOPMed gnomAD |
|
| rs1188842421 | 801 | S>F | No | gnomAD | |
| rs527720055 | 803 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs756820633 | 803 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs753312100 | 804 | P>L | No | ExAC | |
| rs949120910 | 805 | R>C | No |
TOPMed gnomAD |
|
| rs755374301 | 805 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs755374301 | 805 | R>L | No |
ExAC TOPMed gnomAD |
|
|
rs1237537995 COSM1158234 |
806 | R>Q | pancreas [Cosmic] | No |
cosmic curated TOPMed gnomAD |
|
rs754111307 COSM1239399 |
806 | R>W | Variant assessed as Somatic; MODERATE impact. oesophagus [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1437582514 | 807 | S>R | No | TOPMed | |
| rs766645408 | 808 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs2053930729 | 810 | P>A | No |
TOPMed gnomAD |
|
| rs1366047715 | 810 | P>L | No | TOPMed | |
| rs2053930617 | 811 | P>A | No |
TOPMed gnomAD |
|
| rs376367504 | 811 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs376367504 | 811 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs2053930617 | 811 | P>S | No |
TOPMed gnomAD |
|
| rs2053930438 | 812 | S>T | No |
TOPMed gnomAD |
|
| TCGA novel | 813 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761825090 | 813 | R>G | No |
ExAC gnomAD |
|
| rs375769231 | 813 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs991014475 | 815 | L>P | No | gnomAD | |
| rs762832980 | 816 | F>L | No |
ExAC gnomAD |
|
| rs1363212918 | 816 | F>S | No |
TOPMed gnomAD |
|
| rs775202278 | 817 | K>E | No |
ExAC gnomAD |
|
| rs769676505 | 817 | K>R | No |
ExAC gnomAD |
|
| rs1463097239 | 818 | K>E | No |
TOPMed gnomAD |
|
| rs1168740105 | 818 | K>T | No |
TOPMed gnomAD |
|
| rs1301985510 | 819 | E>D | No |
TOPMed gnomAD |
|
| rs2053929752 | 819 | E>G | No | Ensembl | |
| rs2053929808 | 819 | E>K | No | Ensembl | |
| rs2053929631 | 820 | E>D | No | gnomAD | |
| rs2139704550 | 821 | P>T | No | Ensembl | |
|
COSM1579842 rs1454048742 |
822 | M>I | central_nervous_system [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs2053929518 | 822 | M>V | No | Ensembl | |
| rs780975980 | 823 | L>P | No |
ExAC gnomAD |
|
| rs936340633 | 824 | L>S | No |
TOPMed gnomAD |
|
| rs770575759 | 825 | L>R | No |
ExAC gnomAD |
|
| rs2053929181 | 825 | L>V | No | gnomAD | |
| rs1263948044 | 826 | G>E | No | gnomAD | |
| rs1293853910 | 827 | D>Y | No | TOPMed | |
| rs746450605 | 828 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs779504029 | 830 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs1333176009 | 832 | L>P | No | gnomAD | |
| rs755389971 | 833 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1327616313 | 834 | L>P | No | gnomAD | |
| TCGA novel | 835 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1292893278 | 836 | S>F | No | gnomAD | |
| rs2053928132 | 837 | L>F | No |
TOPMed gnomAD |
|
| rs541892601 | 840 | I>M | No |
1000Genomes ExAC gnomAD |
|
| rs767662014 | 840 | I>T | No |
ExAC gnomAD |
|
| rs369776021 | 840 | I>V | No |
ESP ExAC TOPMed |
|
| COSM4819111 | 841 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764215927 | 842 | E>* | No |
ExAC gnomAD |
|
| rs764215927 | 842 | E>K | No |
ExAC gnomAD |
|
| rs2053927414 | 844 | N>H | No | gnomAD | |
| COSM3497580 | 845 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs140360648 COSM175520 |
847 | R>C | large_intestine [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs140360648 | 847 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs775490417 | 847 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs775490417 | 847 | R>L | No |
ExAC gnomAD |
|
| rs1449370458 | 848 | S>C | No | gnomAD | |
| rs769767191 | 851 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs200359887 | 851 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs909851769 | 852 | S>F | No |
TOPMed gnomAD |
|
| rs1289631586 | 852 | S>T | No | gnomAD | |
| rs909851769 | 852 | S>Y | No |
TOPMed gnomAD |
|
| rs2053926764 | 853 | D>G | No | Ensembl | |
| rs986655970 | 853 | D>N | No |
TOPMed gnomAD |
|
| rs199653677 | 854 | S>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs200036746 | 854 | S>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3497578 rs1433904314 |
855 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
|
rs771607431 COSM3497579 |
855 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| COSM3497577 | 856 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs749818618 | 858 | V>L | No |
ExAC gnomAD |
|
| rs780482958 | 859 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2053926177 | 860 | Y>H | No | TOPMed | |
| rs756476859 | 862 | M>I | No |
ExAC gnomAD |
|
| rs146720121 | 862 | M>R | No |
ESP TOPMed gnomAD |
|
| rs146720121 | 862 | M>T | No |
ESP TOPMed gnomAD |
|
| rs2053926064 | 862 | M>V | No | gnomAD | |
| rs542006154 | 863 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs2053925687 | 864 | V>I | No | Ensembl | |
| rs1429141899 | 865 | S>G | No | gnomAD | |
| rs1028400714 | 865 | S>N | No | Ensembl | |
| rs1395770933 | 865 | S>R | No |
TOPMed gnomAD |
|
| rs1195984670 | 866 | P>L | No | gnomAD | |
| rs2053925346 | 867 | V>G | No | Ensembl | |
| rs1477358816 | 867 | V>L | No | gnomAD | |
| rs537793324 | 868 | E>* | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs537793324 | 868 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs148644552 | 869 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1247916454 | 870 | P>R | No |
TOPMed gnomAD |
|
| rs898483258 | 870 | P>T | No | Ensembl | |
| rs1281639165 | 871 | P>L | No | TOPMed | |
| rs955976237 | 871 | P>S | No |
TOPMed gnomAD |
|
| rs2139703685 | 873 | S>G | No | Ensembl | |
| rs1184851386 | 874 | P>L | No | TOPMed | |
| rs200462474 | 875 | C>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs61746639 | 877 | H>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs61746639 | 877 | H>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs3829955 | 878 | N>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs765196740 | 881 | V>F | No |
ExAC gnomAD |
|
| rs1377194661 | 882 | N>D | No | TOPMed | |
| rs1409456795 | 882 | N>S | No |
TOPMed gnomAD |
|
| rs563264687 | 884 | R>* | No | gnomAD | |
| rs563264687 | 884 | R>G | No | gnomAD | |
| rs201816172 | 884 | R>L | No |
1000Genomes ExAC gnomAD |
|
| rs201816172 | 884 | R>P | No |
1000Genomes ExAC gnomAD |
|
| rs201816172 | 884 | R>Q | No |
1000Genomes ExAC gnomAD |
|
| rs776517219 | 885 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs760285196 | 887 | R>C | No |
ExAC TOPMed gnomAD |
|
|
rs772906765 COSM3690173 |
887 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs772906765 | 887 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs760285196 | 887 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs375220969 | 889 | K>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs375220969 | 889 | K>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1202188625 | 890 | R>* | No | gnomAD | |
| rs148435060 | 890 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM3497575 | 892 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 894 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053923005 | 894 | Q>P | No | TOPMed | |
| rs1820763742 | 895 | S>Y | No |
TOPMed gnomAD |
|
| rs746355381 | 896 | L>M | No |
ExAC gnomAD |
|
| rs866235964 | 898 | P>A | No | gnomAD | |
| TCGA novel | 898 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs866235964 | 898 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1441358110 | 902 | T>I | No | gnomAD | |
| rs1284819361 | 903 | L>F | No |
TOPMed gnomAD |
|
| rs2139703358 | 905 | T>A | No | Ensembl | |
| rs779763210 | 905 | T>I | No | gnomAD | |
| rs757632696 | 906 | P>A | No |
ExAC gnomAD |
|
| rs757632696 | 906 | P>S | No |
ExAC gnomAD |
|
| rs200777804 | 907 | S>L | No |
1000Genomes ExAC gnomAD |
|
| rs934157684 | 907 | S>P | No | TOPMed | |
| rs774413853 | 907 | S>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1158230879 | 908 | Q>R | No | Ensembl | |
| rs2053922021 | 909 | P>L | No | TOPMed | |
| rs1364350366 | 909 | P>T | No |
TOPMed gnomAD |
|
| rs200964112 | 910 | S>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs2053921902 | 910 | S>I | No | TOPMed | |
| rs765286426 | 911 | S>N | No |
ExAC gnomAD |
|
| TCGA novel | 911 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs370094241 | 913 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs200148074 | 913 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs371754296 | 914 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs376380612 | 914 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3497573 rs1419510662 |
917 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs142743756 | 918 | D>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs767160217 | 919 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs761253772 | 920 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs2053921172 | 921 | L>P | No | Ensembl | |
| rs2139703061 | 921 | L>V | No | Ensembl | |
| rs1463538851 | 923 | P>R | No | gnomAD | |
| rs777104984 | 924 | E>G | No |
ExAC gnomAD |
|
| rs760106094 | 924 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1594762235 | 925 | T>P | No | Ensembl | |
| rs747328500 | 925 | T>S | No | ExAC | |
| rs1226822540 | 926 | L>F | No | gnomAD | |
| rs1270918759 | 927 | L>R | No | gnomAD | |
| rs2053920658 | 927 | L>V | No | gnomAD | |
| rs2139702918 | 928 | A>G | No | Ensembl | |
| rs777890302 | 929 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs777890302 | 929 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1044774428 | 929 | S>I | No | TOPMed | |
| rs772290301 | 930 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs1462257851 | 932 | P>S | No | gnomAD | |
| rs2053920230 | 933 | S>F | No | TOPMed | |
| rs1594762181 | 934 | S>G | No | Ensembl | |
| rs2053920081 | 935 | N>D | No | gnomAD | |
| rs1162192722 | 935 | N>S | No | gnomAD | |
| rs1468105779 | 936 | G>E | No |
TOPMed gnomAD |
|
| COSM1477737 | 937 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1422938149 | 937 | L>S | No | gnomAD | |
| rs779175250 | 938 | S>G | No |
ExAC gnomAD |
|
| rs1187802997 | 939 | P>L | No | gnomAD | |
| rs948860949 | 940 | S>G | No | TOPMed | |
| TCGA novel | 940 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2053919588 | 941 | P>R | No | Ensembl | |
| rs755104317 | 941 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs755104317 | 941 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs1247283614 | 943 | A>V | No |
TOPMed gnomAD |
|
| rs188647861 | 944 | G>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs188647861 | 944 | G>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1173767110 | 947 | K>E | No | gnomAD | |
| rs1436484925 | 947 | K>T | No | gnomAD | |
| rs147509181 | 948 | T>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs147509181 | 948 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1482976231 | 948 | T>S | No | TOPMed | |
| rs763672830 | 949 | P>A | No |
ExAC gnomAD |
|
| TCGA novel | 949 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762483751 | 949 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1184598561 | 951 | P>L | No |
TOPMed gnomAD |
|
| rs752205589 | 951 | P>T | No |
ExAC gnomAD |
|
| rs267604044 | 952 | S>R | No |
ExAC TOPMed gnomAD |
|
| rs761285831 | 953 | R>* | No |
ExAC TOPMed gnomAD |
|
| rs761285831 | 953 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs773588650 | 953 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs2053890979 | 954 | D>N | No |
TOPMed gnomAD |
|
| rs1484289390 | 956 | G>A | No | gnomAD | |
|
rs2053890878 COSM553596 COSM6076165 |
956 | G>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic gnomAD |
| rs1254727141 | 957 | E>K | No |
TOPMed gnomAD |
|
| COSM433316 | 959 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2053890641 | 959 | P>L | No |
TOPMed gnomAD |
|
| rs767982685 | 960 | R>C | No |
ExAC gnomAD |
|
| rs1290657401 | 960 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2053890398 | 964 | P>S | No | TOPMed | |
| rs138444992 | 965 | N>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1399442138 | 966 | V>M | No | gnomAD | |
| rs2139698317 | 967 | V>G | No | Ensembl | |
| rs1346743038 | 969 | P>S | No | gnomAD | |
| rs775676672 | 970 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs745872449 | 970 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs745872449 | 970 | P>Q | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 970 | P>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775676672 | 970 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs775676672 | 970 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs913263535 | 971 | T>I | No | gnomAD | |
| rs913263535 | 971 | T>N | No | gnomAD | |
| rs780929648 | 974 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs373523963 | 974 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs373523963 | 974 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1200369157 | 975 | W>* | No | gnomAD | |
| rs374751641 | 978 | Q>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs752262464 | 979 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs2053889084 | 980 | D>Y | No | gnomAD | |
| rs1029519642 | 981 | S>P | No |
TOPMed gnomAD |
|
| rs1328946617 | 982 | T>I | No | gnomAD | |
| rs1229737528 | 985 | R>I | No | TOPMed | |
| rs1594760391 | 985 | R>S | No | Ensembl | |
|
rs966033544 COSM6076166 COSM553597 |
990 | E>D | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
cosmic curated NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs202240308 | 990 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs750944190 | 991 | F>L | No |
ExAC gnomAD |
|
| TCGA novel | 992 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs768096501 | 993 | P>A | No |
ExAC gnomAD |
|
|
COSM5456457 rs774902596 |
994 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
|
rs762288591 COSM3356709 |
994 | R>W | Variant assessed as Somatic; MODERATE impact. haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1394945189 | 995 | P>L | No |
TOPMed gnomAD |
|
|
COSM252596 rs1459689257 |
996 | R>C | ovary [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs1167191415 | 996 | R>H | No |
TOPMed gnomAD |
|
| rs1594760323 | 997 | P>A | No | Ensembl | |
| rs763330038 | 998 | S>F | No |
ExAC gnomAD |
|
| rs769927112 | 999 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs769927112 | 999 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs2053887656 | 1000 | N>H | No | Ensembl | |
| rs776665738 | 1000 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs776665738 | 1000 | N>T | No |
ExAC TOPMed gnomAD |
|
| COSM3497572 | 1000 | N>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs746929733 | 1001 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs371313853 | 1001 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs747893572 | 1003 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
|
rs758186971 COSM3690172 |
1003 | R>W | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1594760273 | 1005 | D>A | No | Ensembl | |
|
rs778373003 COSM5534253 |
1006 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2053886697 | 1008 | W>* | No | Ensembl | |
| rs1227456049 | 1008 | W>R | No | gnomAD | |
| rs138921681 | 1010 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs757877089 | 1011 | S>F | No |
ExAC gnomAD |
|
| rs1594760253 | 1011 | S>P | No | Ensembl | |
| rs757877089 | 1011 | S>Y | No |
ExAC gnomAD |
|
| rs1325355223 | 1012 | P>S | No | gnomAD | |
| rs1388788317 | 1013 | S>R | No | gnomAD | |
| rs555763754 | 1014 | H>D | No |
1000Genomes ExAC gnomAD |
|
| rs753034187 | 1014 | H>R | No |
TOPMed gnomAD |
|
| rs766056350 | 1015 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs766056350 | 1015 | A>S | No |
ExAC TOPMed gnomAD |
|
|
rs201827381 COSM1370854 |
1016 | R>C | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1025327866 | 1016 | R>H | No |
TOPMed gnomAD |
|
| rs1253987862 | 1017 | S>C | No | gnomAD | |
| rs1253987862 | 1017 | S>G | No | gnomAD | |
| rs1201529344 | 1017 | S>N | No | gnomAD | |
| rs1594760182 | 1018 | T>P | No | Ensembl | |
| TCGA novel | 1019 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1450260196 | 1020 | P>A | No | gnomAD | |
| rs759687858 | 1020 | P>L | No |
ExAC TOPMed gnomAD |
|
| COSM3420001 | 1020 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1021 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374489590 | 1024 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs959992514 | 1026 | T>A | No | Ensembl | |
| rs2053885056 | 1027 | E>Q | No | gnomAD | |
| rs148083571 | 1028 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs148083571 | 1028 | T>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs771868006 | 1036 | F>L | No |
ExAC gnomAD |
|
| rs180995878 | 1037 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2053884447 | 1038 | S>G | No | TOPMed | |
| rs778649452 | 1039 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs771498795 | 1040 | S>N | No |
ExAC gnomAD |
|
| rs145723754 | 1040 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1243404785 | 1041 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
|
COSM1195797 rs371408609 |
1042 | T>I | lung [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs933532598 | 1043 | V>A | No |
TOPMed gnomAD |
|
| rs933532598 | 1043 | V>G | No |
TOPMed gnomAD |
|
| rs2053883865 | 1044 | E>A | No | TOPMed | |
| rs2053883810 | 1045 | E>* | No | Ensembl | |
| rs2053883810 | 1045 | E>K | No | Ensembl | |
| rs758833277 | 1046 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs778353767 | 1046 | R>W | No |
ExAC TOPMed gnomAD |
|
|
rs752959427 COSM553599 |
1047 | P>R | lung [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs1333730804 | 1048 | G>E | No |
TOPMed gnomAD |
|
| rs1332877920 | 1048 | G>R | No |
TOPMed gnomAD |
|
| rs1483868100 | 1049 | L>F | No | gnomAD | |
| rs1287957693 | 1050 | P>L | No | TOPMed | |
| rs965981059 | 1051 | A>P | No |
TOPMed gnomAD |
|
| rs965981059 | 1051 | A>S | No |
TOPMed gnomAD |
|
| rs1263404910 | 1052 | L>P | No | TOPMed | |
| rs759720980 | 1052 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs201818307 | 1053 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2053882942 | 1053 | L>P | No | Ensembl | |
| rs527635627 | 1054 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs867399337 | 1054 | P>S | No | TOPMed | |
| rs1344022002 | 1055 | F>L | No | gnomAD | |
| rs1217423969 | 1055 | F>L | No |
TOPMed gnomAD |
|
| rs1301363942 | 1056 | Q>E | No | gnomAD | |
| rs983813911 | 1056 | Q>R | No | TOPMed | |
| rs773139386 | 1058 | G>E | No |
ExAC gnomAD |
|
| rs1279992477 | 1058 | G>R | No |
TOPMed gnomAD |
|
| rs146739032 | 1059 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs768491592 | 1060 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2053882095 | 1061 | P>L | No |
TOPMed gnomAD |
|
| rs749003418 | 1062 | P>L | No |
ExAC TOPMed gnomAD |
|
| COSM3497571 | 1062 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs560713687 | 1063 | T>A | No |
1000Genomes ExAC gnomAD |
|
| rs1294610686 | 1063 | T>N | No | Ensembl | |
| rs560713687 | 1063 | T>P | No |
1000Genomes ExAC gnomAD |
|
| rs560713687 | 1063 | T>S | No |
1000Genomes ExAC gnomAD |
|
| rs778449870 | 1064 | E>A | No |
ExAC gnomAD |
|
| rs745327663 | 1064 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs745327663 | 1064 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs778449870 | 1064 | E>V | No |
ExAC gnomAD |
|
| rs374670001 | 1065 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs190369113 | 1065 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779213783 | 1066 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2053881387 | 1067 | L>P | No | TOPMed | |
| rs968520584 | 1069 | D>E | No | TOPMed | |
| rs1310892350 | 1069 | D>Y | No |
TOPMed gnomAD |
|
| rs1594759855 | 1072 | A>V | No | Ensembl | |
| rs750551594 | 1074 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs1219637132 | 1075 | Q>* | No | gnomAD | |
| rs761721768 | 1079 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs201830876 | 1081 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs201830876 | 1081 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs775309790 COSM957418 |
1082 | P>L | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1286093311 | 1082 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs769526586 | 1083 | L>Q | No |
ExAC gnomAD |
|
| rs769526586 | 1083 | L>R | No |
ExAC gnomAD |
|
| TCGA novel | 1084 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs776047235 | 1086 | A>G | No |
ExAC TOPMed gnomAD |
|
| rs1386927844 | 1086 | A>S | No |
TOPMed gnomAD |
|
|
COSM4700412 rs776047235 |
1086 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs748663344 | 1087 | E>G | No |
ExAC gnomAD |
|
| rs1307993783 | 1088 | L>P | No |
TOPMed gnomAD |
|
| rs755302161 | 1088 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1411795797 | 1090 | T>K | No |
TOPMed gnomAD |
|
| rs1248528644 | 1091 | H>Q | No |
TOPMed gnomAD |
|
| rs1190436703 | 1092 | R>K | No | gnomAD | |
| rs749614315 | 1093 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs780374496 | 1093 | P>H | No |
ExAC TOPMed gnomAD |
|
|
rs780374496 COSM2136247 |
1093 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs749614315 | 1093 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs756368396 | 1095 | P>A | No |
ExAC gnomAD |
|
| rs1322590535 | 1095 | P>L | No | gnomAD | |
| rs887919504 | 1096 | Y>* | No |
TOPMed gnomAD |
|
| rs1396834166 | 1096 | Y>C | No | TOPMed | |
| rs1396834166 | 1096 | Y>F | No | TOPMed | |
| rs1355739175 | 1097 | E>G | No | gnomAD | |
| rs750637587 | 1098 | I>S | No |
ExAC gnomAD |
|
| rs750637587 | 1098 | I>T | No |
ExAC gnomAD |
|
| rs1047892994 | 1102 | F>C | No | Ensembl | |
| rs757301027 | 1104 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs1566725269 | 1105 | S>W | No | Ensembl |
1 associated diseases with P80192
Without disease ID
5 regional properties for P80192
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 11 - 118 | IPR000980 |
| domain | SH3 domain | 132 - 192 | IPR001452-1 |
| domain | SH3 domain | 235 - 296 | IPR001452-2 |
| domain | CRK, N-terminal SH3 domain | 135 - 189 | IPR035457 |
| domain | CRK, C-terminal SH3 domain | 237 - 293 | IPR035458 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.- | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| JUN kinase kinase kinase activity | Catalysis of the reaction |
| MAP kinase kinase activity | Catalysis of the concomitant phosphorylation of threonine (T) and tyrosine (Y) residues in a Thr-Glu-Tyr (TEY) thiolester sequence in a MAP kinase (MAPK) substrate. |
| molecular function activator activity | A molecular function regulator that activates or increases the activity of its target via non-covalent binding that does not result in covalent modification to the target. |
| protein homodimerization activity | Binding to an identical protein to form a homodimer. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
41 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| A2VDU3 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Bos taurus (Bovine) | SS |
| Q4TVR5 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Bos taurus (Bovine) | PR |
| Q3SZJ2 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Bos taurus (Bovine) | PR |
| Q6XUX0 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Gallus gallus (Chicken) | PR |
| P83104 | Takl1 | Putative mitogen-activated protein kinase kinase kinase 7-like | Drosophila melanogaster (Fruit fly) | PR |
| Q95UN8 | slpr | Mitogen-activated protein kinase kinase kinase | Drosophila melanogaster (Fruit fly) | EV |
| P00540 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Homo sapiens (Human) | PR |
| Q5TCX8 | MAP3K21 | Mitogen-activated protein kinase kinase kinase 21 | Homo sapiens (Human) | PR |
| Q6XUX3 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Homo sapiens (Human) | PR |
| O43318 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Homo sapiens (Human) | SS |
| Q9NYL2 | MAP3K20 | Mitogen-activated protein kinase kinase kinase 20 | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| O43353 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Homo sapiens (Human) | PR |
| Q02779 | MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | Homo sapiens (Human) | SS |
| Q16584 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | Homo sapiens (Human) | EV |
| Q13418 | ILK | Integrin-linked protein kinase | Homo sapiens (Human) | PR |
| P04049 | RAF1 | RAF proto-oncogene serine/threonine-protein kinase | Homo sapiens (Human) | EV |
| P10398 | ARAF | Serine/threonine-protein kinase A-Raf | Homo sapiens (Human) | PR |
| P15056 | BRAF | Serine/threonine-protein kinase B-raf | Homo sapiens (Human) | EV |
| Q8NB16 | MLKL | Mixed lineage kinase domain-like protein | Homo sapiens (Human) | EV |
| P58801 | Ripk2 | Receptor-interacting serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q62073 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Mus musculus (Mouse) | EV |
| Q9D2Y4 | Mlkl | Mixed lineage kinase domain-like protein | Mus musculus (Mouse) | SS |
| Q9ESL4 | Map3k20 | Mitogen-activated protein kinase kinase kinase 20 | Mus musculus (Mouse) | PR |
| P00536 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Mus musculus (Mouse) | PR |
| Q66L42 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Mus musculus (Mouse) | SS |
| Q80XI6 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Mus musculus (Mouse) | PR |
| Q8VDG6 | Map3k21 | Mitogen-activated protein kinase kinase kinase 21 | Mus musculus (Mouse) | PR |
| Q3U1V8 | Map3k9 | Mitogen-activated protein kinase kinase kinase 9 | Mus musculus (Mouse) | SS |
| P0C8E4 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Rattus norvegicus (Rat) | SS |
| D3ZG83 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Rattus norvegicus (Rat) | SS |
| P00539 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Rattus norvegicus (Rat) | PR |
| Q66HA1 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Rattus norvegicus (Rat) | PR |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9FPR3 | EDR1 | Serine/threonine-protein kinase EDR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q05609 | CTR1 | Serine/threonine-protein kinase CTR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q67E00 | dstyk | Dual serine/threonine and tyrosine protein kinase | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MEPSRALLGC | LASAAAAAPP | GEDGAGAGAE | EEEEEEEEAA | AAVGPGELGC | DAPLPYWTAV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| FEYEAAGEDE | LTLRLGDVVE | VLSKDSQVSG | DEGWWTGQLN | QRVGIFPSNY | VTPRSAFSSR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| CQPGGEDPSC | YPPIQLLEID | FAELTLEEII | GIGGFGKVYR | AFWIGDEVAV | KAARHDPDED |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ISQTIENVRQ | EAKLFAMLKH | PNIIALRGVC | LKEPNLCLVM | EFARGGPLNR | VLSGKRIPPD |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ILVNWAVQIA | RGMNYLHDEA | IVPIIHRDLK | SSNILILQKV | ENGDLSNKIL | KITDFGLARE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| WHRTTKMSAA | GTYAWMAPEV | IRASMFSKGS | DVWSYGVLLW | ELLTGEVPFR | GIDGLAVAYG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| VAMNKLALPI | PSTCPEPFAK | LMEDCWNPDP | HSRPSFTNIL | DQLTTIEESG | FFEMPKDSFH |
| 430 | 440 | 450 | 460 | 470 | 480 |
| CLQDNWKHEI | QEMFDQLRAK | EKELRTWEEE | LTRAALQQKN | QEELLRRREQ | ELAEREIDIL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| ERELNIIIHQ | LCQEKPRVKK | RKGKFRKSRL | KLKDGNRISL | PSDFQHKFTV | QASPTMDKRK |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SLINSRSSPP | ASPTIIPRLR | AIQLTPGESS | KTWGRSSVVP | KEEGEEEEKR | APKKKGRTWG |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PGTLGQKELA | SGDEGSPQRR | EKANGLSTPS | ESPHFHLGLK | SLVDGYKQWS | SSAPNLVKGP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RSSPALPGFT | SLMEMEDEDS | EGPGSGESRL | QHSPSQSYLC | IPFPRGEDGD | GPSSDGIHEE |
| 730 | 740 | 750 | 760 | 770 | 780 |
| PTPVNSATST | PQLTPTNSLK | RGGAHHRRCE | VALLGCGAVL | AATGLGFDLL | EAGKCQLLPL |
| 790 | 800 | 810 | 820 | 830 | 840 |
| EEPEPPAREE | KKRREGLFQR | SSRPRRSTSP | PSRKLFKKEE | PMLLLGDPSA | SLTLLSLSSI |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SECNSTRSLL | RSDSDEIVVY | EMPVSPVEAP | PLSPCTHNPL | VNVRVERFKR | DPNQSLTPTH |
| 910 | 920 | 930 | 940 | 950 | 960 |
| VTLTTPSQPS | SHRRTPSDGA | LKPETLLASR | SPSSNGLSPS | PGAGMLKTPS | PSRDPGEFPR |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LPDPNVVFPP | TPRRWNTQQD | STLERPKTLE | FLPRPRPSAN | RQRLDPWWFV | SPSHARSTSP |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| ANSSSTETPS | NLDSCFASSS | STVEERPGLP | ALLPFQAGPL | PPTERTLLDL | DAEGQSQDST |
| 1090 | 1100 | ||||
| VPLCRAELNT | HRPAPYEIQQ | EFWS |