P67998
Gene name |
RPS6KB1 |
Protein name |
Ribosomal protein S6 kinase beta-1 |
Names |
S6K-beta-1, S6K1, 70 kDa ribosomal protein S6 kinase 1, P70S6K1, p70-S6K 1, Ribosomal protein S6 kinase I, p70 ribosomal S6 kinase alpha, p70 S6 kinase alpha, p70 S6K-alpha, p70 S6KA |
Species |
Oryctolagus cuniculus (Rabbit) |
KEGG Pathway |
ocu:100009260 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
91-352 (Protein kinase domain) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
235-258 (Activation loop from InterPro)
Target domain |
91-352 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Lai KO et al. (2015) "Cyclin-dependent Kinase 5 (Cdk5)-dependent Phosphorylation of p70 Ribosomal S6 Kinase 1 (S6K) Is Required for Dendritic Spine Morphogenesis", The Journal of biological chemistry, 290, 14637-46
- Dennis PB et al. (1998) "Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation", The Journal of biological chemistry, 273, 14845-52
- Mahalingam M et al. (1996) "Constitutive activation of S6 kinase by deletion of amino-terminal autoinhibitory and rapamycin sensitivity domains", Molecular and cellular biology, 16, 405-13
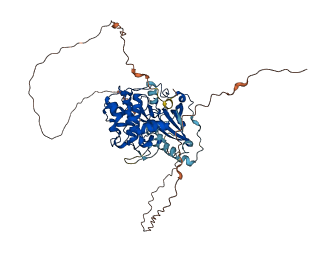
Autoinhibited structure

Activated structure

1 structures for P67998
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P67998-F1 | Predicted | AlphaFoldDB |
No variants for P67998
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for P67998 | |||||
No associated diseases with P67998
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| synapse | The junction between an axon of one neuron and a dendrite of another neuron, a muscle fiber or a glial cell. As the axon approaches the synapse it enlarges into a specialized structure, the presynaptic terminal bouton, which contains mitochondria and synaptic vesicles. At the tip of the terminal bouton is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic terminal bouton secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cellular response to growth factor stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a growth factor stimulus. |
| long-chain fatty acid import into cell | The directed movement of a long-chain fatty acid from outside of a cell into a cell. This may occur via transport across the plasma membrane or via endocytosis. A long-chain fatty acid is a fatty acid with a chain length between C13 and C22. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of insulin receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of insulin receptor signaling. |
| peptidyl-serine phosphorylation | The phosphorylation of peptidyl-serine to form peptidyl-O-phospho-L-serine. |
| TOR signaling | The series of molecular signals mediated by TOR (Target of rapamycin) proteins, members of the phosphoinositide (PI) 3-kinase related kinase (PIKK) family that act as serine/threonine kinases in response to nutrient availability or growth factors. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRRRRRRDGF | YPAPDFRDRE | AEDMAGVFDI | DLDQPEDAGS | EDELEEGGQL | NESMDHGGVG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PYELGMEHCE | KFEISETSVN | RGPEKIRPEC | FELLRVLGKG | GYGKVFQVRK | VTGANTGKIF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AMKVLKKAMI | VRNAKDTAHT | KAERNILEEV | KHPFIVDLIY | AFQTGGKLYL | ILEYLSGGEL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| FMQLEREGIF | MEDTACFYLA | EISMALGHLH | QKGIIYRDLK | PENIMLNHQG | HVKLTDFGLC |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KESIHDGTVT | HTFCGTIEYM | APEILMRSGH | NRAVDWWSLG | ALMYDMLTGA | PPFTGENRKK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| TIDKILKCKL | NLPPYLTQEA | RDLLKKLLKR | NAASRLGAGP | GDAGEVQAHP | FFRHINWEEL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LARKVEPPFK | PLLQSEEDVS | QFDSKFTRQT | PVDSPDDSTL | SESANQVFLG | FTYVAPSVLE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| SVKEKFSFEP | KIRSPRRFIG | SPRTPVSPVK | FSPGDFWGRG | ASASTANPQT | PVEYPMETSG |
| 490 | 500 | 510 | 520 | ||
| IEQMDVTTSG | EASAPLPIRQ | PNSGPYKKQA | FPMISKRPEH | LRMNL |