P63096
Gene name |
GNAI1 |
Protein name |
Guanine nucleotide-binding protein G |
Names |
i subunit alpha-1 , Adenylate cyclase-inhibiting G alpha protein |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:2770 |
EC number |
|
Protein Class |
GTP-BINDING PROTEIN ALPHA SUBUNIT (PTHR10218) |
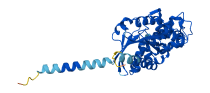
Descriptions
Gαi1 is a G-protein alpha subunit that plays a crucial role in signal transduction pathway initiated by G-protein-coupled receptor (GPCR) activation. G proteins function as transducers downstream of GPCRs in numerous signaling cascades. The alpha subunit contains the guanine nucleotide binding site and alternates between an active (GTP-bound state) and an inactive (GDP-bound state). Activated GPCR induces conformational change in Gαi1, which allows for GDP-to-GTP exchange by binding to the GPCR-binding site within a Ras-like domain, leading to dissociation of the Gαi1 subunit and initiation of downstream signaling.
Autoinhibitory domains (AIDs)
Target domain |
184-348 (Ras-like domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
Structural analysis, Deletion assay |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

296 structures for P63096
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1KJY | X-ray | 270 A | A/C | 30-354 | PDB |
| 1Y3A | X-ray | 250 A | A/B/C/D | 26-354 | PDB |
| 2G83 | X-ray | 280 A | A/B | 33-345 | PDB |
| 2GTP | X-ray | 255 A | A/B | 32-354 | PDB |
| 2IK8 | X-ray | 271 A | A/C | 31-354 | PDB |
| 2OM2 | X-ray | 220 A | A/C | 31-354 | PDB |
| 2XNS | X-ray | 341 A | A/B | 30-354 | PDB |
| 3ONW | X-ray | 238 A | A/B | 31-354 | PDB |
| 3QE0 | X-ray | 300 A | A/B/C | 33-354 | PDB |
| 3QI2 | X-ray | 280 A | A/B | 31-354 | PDB |
| 3UMR | X-ray | 224 A | A | 1-354 | PDB |
| 3UMS | X-ray | 260 A | A | 1-354 | PDB |
| 4G5Q | X-ray | 290 A | A/B/C/D | 25-354 | PDB |
| 5JS7 | NMR | - | A | 30-354 | PDB |
| 5JS8 | NMR | - | A | 30-354 | PDB |
| 5TDH | X-ray | 300 A | A/H | 1-354 | PDB |
| 6CMO | EM | 450 A | A | 1-354 | PDB |
| 6CRK | X-ray | 200 A | A | 2-354 | PDB |
| 6DDE | EM | 350 A | A | 1-354 | PDB |
| 6DDF | EM | 350 A | A | 1-354 | PDB |
| 6KPF | EM | 290 A | A | 1-354 | PDB |
| 6KPG | EM | 300 A | A | 2-354 | PDB |
| 6LFM | EM | 350 A | A | 2-354 | PDB |
| 6LFO | EM | 340 A | A | 2-354 | PDB |
| 6LML | EM | 390 A | A | 1-354 | PDB |
| 6N4B | EM | 300 A | A | 1-354 | PDB |
| 6OMM | EM | 317 A | A | 2-354 | PDB |
| 6OS9 | EM | 300 A | A | 1-354 | PDB |
| 6OSA | EM | 300 A | A | 1-354 | PDB |
| 6OT0 | EM | 390 A | A | 1-354 | PDB |
| 6PB0 | EM | 300 A | A | 61-184 | PDB |
| 6PB1 | EM | 280 A | A | 61-184 | PDB |
| 6PT0 | EM | 320 A | A | 1-354 | PDB |
| 6QNO | EM | 438 A | A | 1-354 | PDB |
| 6VU8 | EM | 414 A | B | 2-354 | PDB |
| 6XBJ | EM | 388 A | A | 1-354 | PDB |
| 6XBK | EM | 324 A | A | 1-354 | PDB |
| 6XBL | EM | 390 A | A | 1-354 | PDB |
| 6XBM | EM | 315 A | A | 1-354 | PDB |
| 7CMU | EM | 300 A | A | 1-354 | PDB |
| 7CMV | EM | 270 A | A | 1-354 | PDB |
| 7DB6 | EM | 330 A | A | 1-354 | PDB |
| 7DW9 | EM | 260 A | PDB | ||
| 7E2X | EM | 300 A | A | 1-354 | PDB |
| 7E2Y | EM | 300 A | A | 1-354 | PDB |
| 7E2Z | EM | 310 A | A | 1-354 | PDB |
| 7E32 | EM | 290 A | A | 1-354 | PDB |
| 7E33 | EM | 290 A | A | 1-354 | PDB |
| 7E9G | EM | 350 A | A | 1-354 | PDB |
| 7EB2 | EM | 350 A | A | 1-354 | PDB |
| 7EJX | EM | 240 A | A | 1-354 | PDB |
| 7EO2 | EM | 289 A | B | 1-354 | PDB |
| 7EO4 | EM | 286 A | B | 1-354 | PDB |
| 7EUO | EM | 290 A | A | 1-354 | PDB |
| 7EVY | EM | 298 A | A | 1-354 | PDB |
| 7EVZ | EM | 307 A | A | 1-354 | PDB |
| 7EW0 | EM | 342 A | A | 1-354 | PDB |
| 7EW1 | EM | 340 A | D | 1-354 | PDB |
| 7EW2 | EM | 310 A | A | 1-354 | PDB |
| 7EW3 | EM | 310 A | A | 1-354 | PDB |
| 7EW4 | EM | 320 A | A | 1-354 | PDB |
| 7EW7 | EM | 327 A | A | 1-354 | PDB |
| 7EXD | EM | 340 A | A | 1-354 | PDB |
| 7EZH | EM | 320 A | A | 1-354 | PDB |
| 7EZK | EM | 310 A | PDB | ||
| 7EZM | EM | 290 A | A | 2-30 | PDB |
| 7F1Q | EM | 290 A | A | 1-354 | PDB |
| 7F1R | EM | 300 A | A | 1-354 | PDB |
| 7F1S | EM | 280 A | A | 1-354 | PDB |
| 7F4D | EM | 300 A | PDB | ||
| 7F4F | EM | 290 A | PDB | ||
| 7F4H | EM | 270 A | PDB | ||
| 7F4I | EM | 310 A | PDB | ||
| 7JHJ | EM | 320 A | A | 2-354 | PDB |
| 7JVR | EM | 280 A | A | 1-354 | PDB |
| 7L0P | EM | 410 A | A | 1-354 | PDB |
| 7L0Q | EM | 430 A | A | 1-354 | PDB |
| 7L0R | EM | 420 A | A | 1-354 | PDB |
| 7L0S | EM | 450 A | A | 1-354 | PDB |
| 7MTS | EM | 320 A | C | 1-354 | PDB |
| 7NA7 | EM | 270 A | A | 1-354 | PDB |
| 7NA8 | EM | 270 A | A | 1-354 | PDB |
| 7O7F | EM | 315 A | A | 1-354 | PDB |
| 7P02 | EM | 287 A | A | 229-242 | PDB |
| 7RKM | EM | 350 A | A | 2-354 | PDB |
| 7RKN | EM | 360 A | A | 2-354 | PDB |
| 7RKX | EM | 310 A | A | 2-354 | PDB |
| 7RKY | EM | 380 A | A | 2-354 | PDB |
| 7S8M | EM | 254 A | B | 1-354 | PDB |
| 7S8O | EM | 258 A | B | 1-354 | PDB |
| 7SBF | EM | 290 A | A | 1-354 | PDB |
| 7SCG | EM | 300 A | A | 1-354 | PDB |
| 7SQO | EM | 317 A | A | 1-354 | PDB |
| 7T2G | EM | 250 A | A | 1-354 | PDB |
| 7T2H | EM | 320 A | A | 1-354 | PDB |
| 7T6B | EM | 319 A | A | 1-32 | PDB |
| 7T6S | EM | 300 A | A | 2-354 | PDB |
| 7T6T | EM | 320 A | A | 2-354 | PDB |
| 7T6U | EM | 290 A | A | 2-354 | PDB |
| 7T6V | EM | 310 A | A | 2-354 | PDB |
| 7TRK | EM | 280 A | A | 1-354 | PDB |
| 7TRP | EM | 240 A | A | 1-354 | PDB |
| 7TRQ | EM | 250 A | A | 1-354 | PDB |
| 7TRS | EM | 280 A | A | 1-354 | PDB |
| 7TUZ | EM | 312 A | A | 1-354 | PDB |
| 7U2K | EM | 330 A | A | 1-354 | PDB |
| 7U2L | EM | 320 A | A | 1-354 | PDB |
| 7V68 | EM | 340 A | A | 1-354 | PDB |
| 7V69 | EM | 340 A | A | 1-354 | PDB |
| 7V6A | EM | 360 A | A | 1-354 | PDB |
| 7V9L | EM | 260 A | PDB | ||
| 7VAB | EM | 320 A | PDB | ||
| 7VDH | EM | 290 A | A | 1-354 | PDB |
| 7VDL | EM | 322 A | A | 1-354 | PDB |
| 7VDM | EM | 298 A | A | 1-354 | PDB |
| 7VFX | EM | 280 A | A | 1-354 | PDB |
| 7VGX | EM | 320 A | A | 2-354 | PDB |
| 7VGY | EM | 310 A | B | 2-354 | PDB |
| 7VGZ | EM | 330 A | C | 2-354 | PDB |
| 7VH0 | EM | 346 A | B | 2-354 | PDB |
| 7VIE | EM | 286 A | D | 1-354 | PDB |
| 7VIF | EM | 283 A | D | 1-354 | PDB |
| 7VIG | EM | 289 A | D | 1-354 | PDB |
| 7VIH | EM | 298 A | D | 1-354 | PDB |
| 7VKT | EM | 290 A | B | 1-354 | PDB |
| 7VL8 | EM | 290 A | A | 1-354 | PDB |
| 7VL9 | EM | 260 A | A | 1-354 | PDB |
| 7VLA | EM | 270 A | A | 1-354 | PDB |
| 7VUG | EM | 320 A | A | 2-354 | PDB |
| 7VUY | EM | 284 A | A | 1-354 | PDB |
| 7VUZ | EM | 289 A | A | 1-354 | PDB |
| 7VV3 | EM | 297 A | A | 1-354 | PDB |
| 7VV5 | EM | 276 A | A | 1-354 | PDB |
| 7W0L | EM | 357 A | A | 1-354 | PDB |
| 7W0M | EM | 371 A | A | 1-354 | PDB |
| 7W0N | EM | 421 A | A | 1-354 | PDB |
| 7W0O | EM | 378 A | A | 1-354 | PDB |
| 7W0P | EM | 316 A | A | 1-354 | PDB |
| 7WF7 | EM | 340 A | B | 1-354 | PDB |
| 7WIC | EM | 280 A | A | 1-354 | PDB |
| 7WIG | EM | 270 A | A | 1-354 | PDB |
| 7WJ5 | EM | 372 A | A | 1-354 | PDB |
| 7WQ3 | EM | 270 A | A | 1-354 | PDB |
| 7WU4 | EM | 340 A | A | 181-354 | PDB |
| 7WU5 | EM | 300 A | PDB | ||
| 7WU9 | EM | 338 A | A | 2-354 | PDB |
| 7WUI | EM | 310 A | PDB | ||
| 7WUJ | EM | 330 A | PDB | ||
| 7WVU | EM | 330 A | A | 1-354 | PDB |
| 7WYB | EM | 297 A | B | 1-354 | PDB |
| 7WZ4 | EM | 300 A | A | 1-354 | PDB |
| 7X2V | EM | 309 A | B | 1-354 | PDB |
| 7X5H | EM | 310 A | A | 1-354 | PDB |
| 7X9A | EM | 320 A | A | 1-354 | PDB |
| 7X9B | EM | 340 A | A | 1-354 | PDB |
| 7X9C | EM | 300 A | A | 1-354 | PDB |
| 7X9Y | EM | 310 A | A | 1-354 | PDB |
| 7XA3 | EM | 290 A | A | 1-354 | PDB |
| 7XAT | EM | 285 A | B | 1-354 | PDB |
| 7XAU | EM | 297 A | B | 1-354 | PDB |
| 7XAV | EM | 287 A | B | 1-354 | PDB |
| 7XBW | EM | 280 A | C | 1-354 | PDB |
| 7XBX | EM | 340 A | C | 1-354 | PDB |
| 7XK2 | EM | 310 A | A | 1-354 | PDB |
| 7XK8 | EM | 330 A | A | 1-354 | PDB |
| 7XMR | EM | 310 A | A | 1-354 | PDB |
| 7XMS | EM | 290 A | A | 1-354 | PDB |
| 7XMT | EM | 280 A | A | 1-354 | PDB |
| 7XTA | EM | 320 A | A | 1-354 | PDB |
| 7XXH | EM | 290 A | A | 2-24 | PDB |
| 7Y12 | EM | 310 A | A | 1-354 | PDB |
| 7Y15 | EM | 290 A | A | 1-354 | PDB |
| 7Y1F | EM | 330 A | A | 1-354 | PDB |
| 7Y64 | EM | 290 A | A | 1-354 | PDB |
| 7Y65 | EM | 320 A | A | 1-354 | PDB |
| 7Y66 | EM | 290 A | A | 1-354 | PDB |
| 7Y67 | EM | 280 A | A | 1-354 | PDB |
| 7Y89 | EM | 302 A | C | 4-354 | PDB |
| 7YAC | EM | 324 A | A | 1-354 | PDB |
| 7YAE | EM | 337 A | A | 1-354 | PDB |
| 7YDJ | EM | 303 A | PDB | ||
| 7YDP | EM | 310 A | PDB | ||
| 7YKD | EM | 281 A | C | 4-354 | PDB |
| 7YON | EM | 295 A | A | 1-354 | PDB |
| 7YOO | EM | 311 A | A | 1-354 | PDB |
| 7YU3 | EM | 340 A | A | 1-354 | PDB |
| 7YU5 | EM | 330 A | A | 1-354 | PDB |
| 7YU6 | EM | 350 A | A | 1-354 | PDB |
| 7YU7 | EM | 380 A | A | 1-354 | PDB |
| 7YU8 | EM | 450 A | A | 1-354 | PDB |
| 8DZP | EM | 271 A | B | 1-354 | PDB |
| 8EF5 | EM | 330 A | A/F | 1-354 | PDB |
| 8EF6 | EM | 320 A | A/F | 1-354 | PDB |
| 8EFB | EM | 320 A | A | 1-354 | PDB |
| 8EFL | EM | 320 A | A/F | 1-354 | PDB |
| 8EFO | EM | 280 A | A/F | 1-354 | PDB |
| 8EFQ | EM | 330 A | A | 1-354 | PDB |
| 8F7Q | EM | 322 A | A/F | 1-354 | PDB |
| 8F7R | EM | 328 A | A/F | 1-354 | PDB |
| 8F7S | EM | 300 A | A/F | 1-354 | PDB |
| 8F7W | EM | 319 A | A | 1-354 | PDB |
| 8F7X | EM | 328 A | A | 1-354 | PDB |
| 8FEG | EM | 254 A | C | 1-354 | PDB |
| 8G05 | EM | 300 A | A | 1-354 | PDB |
| 8G59 | EM | 264 A | A | 1-329 | PDB |
| 8G94 | EM | 315 A | B | 1-354 | PDB |
| 8GCM | EM | 350 A | A | 1-354 | PDB |
| 8GCP | EM | 310 A | A | 1-354 | PDB |
| 8GDC | EM | 350 A | A | 1-354 | PDB |
| 8GHV | EM | 280 A | A | 1-354 | PDB |
| 8GUQ | EM | 308 A | A | 1-354 | PDB |
| 8GUR | EM | 284 A | A | 1-354 | PDB |
| 8GUS | EM | 297 A | A | 1-354 | PDB |
| 8GUT | EM | 298 A | A | 1-354 | PDB |
| 8H2G | EM | 301 A | B | 1-354 | PDB |
| 8H4I | EM | 306 A | PDB | ||
| 8HBD | EM | 299 A | A | 1-354 | PDB |
| 8HK2 | EM | 290 A | B | 2-354 | PDB |
| 8HK3 | EM | 320 A | A | 2-354 | PDB |
| 8HK5 | EM | 300 A | C | 2-354 | PDB |
| 8HN8 | EM | 300 A | A | 1-354 | PDB |
| 8HNK | EM | 301 A | A | 1-354 | PDB |
| 8HNL | EM | 298 A | A | 1-354 | PDB |
| 8HNM | EM | 294 A | A | 1-354 | PDB |
| 8HOC | EM | 300 A | A | 1-354 | PDB |
| 8HQE | EM | 297 A | A | 1-354 | PDB |
| 8HQM | EM | 295 A | A | 1-354 | PDB |
| 8HQN | EM | 300 A | A | 1-354 | PDB |
| 8HS3 | EM | 314 A | A | 1-354 | PDB |
| 8HSC | EM | 322 A | A | 1-354 | PDB |
| 8HVI | EM | 304 A | B | 1-354 | PDB |
| 8I7V | EM | 277 A | B | 1-354 | PDB |
| 8I7W | EM | 339 A | B | 1-354 | PDB |
| 8IA8 | EM | 286 A | C | 4-354 | PDB |
| 8IC0 | EM | 341 A | B | 1-354 | PDB |
| 8ID3 | EM | 310 A | A | 1-354 | PDB |
| 8ID4 | EM | 310 A | A | 1-354 | PDB |
| 8ID6 | EM | 280 A | A | 1-354 | PDB |
| 8ID8 | EM | 300 A | A | 1-354 | PDB |
| 8ID9 | EM | 300 A | A | 1-354 | PDB |
| 8IHB | EM | 285 A | A | 1-354 | PDB |
| 8IHF | EM | 297 A | A | 1-354 | PDB |
| 8IHH | EM | 306 A | A | 1-354 | PDB |
| 8IHI | EM | 311 A | A | 1-354 | PDB |
| 8IHJ | EM | 307 A | A | 1-354 | PDB |
| 8IJ3 | EM | 328 A | C | 4-354 | PDB |
| 8IJA | EM | 269 A | C | 4-354 | PDB |
| 8IJB | EM | 323 A | C | 4-354 | PDB |
| 8IJD | EM | 325 A | C | 4-354 | PDB |
| 8INR | EM | 273 A | PDB | ||
| 8IOC | EM | 286 A | PDB | ||
| 8IOD | EM | 259 A | PDB | ||
| 8IRS | EM | 300 A | A | 1-354 | PDB |
| 8IRT | EM | 270 A | A | 1-354 | PDB |
| 8IRV | EM | 310 A | PDB | ||
| 8IW4 | EM | 349 A | PDB | ||
| 8IW7 | EM | 297 A | PDB | ||
| 8IW9 | EM | 308 A | PDB | ||
| 8IY5 | EM | 280 A | A | 1-354 | PDB |
| 8J18 | EM | 289 A | A | 1-354 | PDB |
| 8J19 | EM | 323 A | A | 1-354 | PDB |
| 8J1A | EM | 324 A | A | 1-354 | PDB |
| 8J20 | EM | 320 A | C | 1-354 | PDB |
| 8J21 | EM | 330 A | C | 1-354 | PDB |
| 8J22 | EM | 320 A | B | 1-354 | PDB |
| 8J24 | EM | 260 A | C | 1-354 | PDB |
| 8J6J | EM | 280 A | A | 1-354 | PDB |
| 8J6P | EM | 255 A | A | 3-354 | PDB |
| 8J6Q | EM | 260 A | A | 3-354 | PDB |
| 8J6R | EM | 276 A | A | 3-354 | PDB |
| 8JD3 | EM | 330 A | A | 1-354 | PDB |
| 8JD5 | EM | 360 A | A | 1-354 | PDB |
| 8JGB | EM | 284 A | A | 1-354 | PDB |
| 8JGG | EM | 300 A | A | 1-354 | PDB |
| 8JHY | EM | 287 A | D | 1-354 | PDB |
| 8JII | EM | 317 A | D | 1-354 | PDB |
| 8JIL | EM | 350 A | D | 1-354 | PDB |
| 8JIM | EM | 298 A | D | 1-354 | PDB |
| 8JIP | EM | 285 A | PDB | ||
| 8JIR | EM | 257 A | PDB | ||
| 8JR9 | EM | 257 A | PDB | ||
| 8JSP | EM | 365 A | A | 4-354 | PDB |
| 8JZ7 | EM | 260 A | D | 1-354 | PDB |
| 8K2X | EM | 320 A | A | 1-354 | PDB |
| 8K4N | EM | 283 A | A | 1-354 | PDB |
| 8KFX | EM | 296 A | A | 1-354 | PDB |
| 8KFY | EM | 306 A | A | 1-354 | PDB |
| 8KFZ | EM | 330 A | A | 1-354 | PDB |
| 8SAI | EM | 327 A | D | 1-354 | PDB |
| 8SG1 | EM | 294 A | A | 4-354 | PDB |
| 8U1U | EM | 310 A | B | 1-354 | PDB |
| 8W8B | EM | 300 A | A | 1-354 | PDB |
| 8W8R | EM | 330 A | A | 1-349 | PDB |
| 8WRB | EM | 291 A | A | 1-354 | PDB |
| 8XBH | EM | 283 A | A | 1-354 | PDB |
| AF-P63096-F1 | Predicted | AlphaFoldDB |
290 variants for P63096
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV002249061 VAR_087204 rs2116052322 |
40 | G>C | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities NEDHISB [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
| VAR_087205 | 40 | G>R | NEDHISB [UniProt] | Yes | UniProt |
| VAR_087206 | 45 | G>D | NEDHISB [UniProt] | Yes | UniProt |
|
rs2115631937 RCV001823048 |
45 | G>V | GNAI1-related disorder [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
| VAR_087207 | 48 | T>I | NEDHISB [UniProt] | Yes | UniProt |
|
RCV002249684 RCV001095673 VAR_087208 rs1788434338 |
48 | T>K | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities Neurodevelopmental disorder NEDHISB [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
rs1788434582 RCV001268270 VAR_087209 |
52 | Q>P | NEDHISB; loss of GTP binding [UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
| VAR_087210 | 75 | S>del | NEDHISB; uncertain significance [UniProt] | Yes | UniProt |
|
RCV001780011 rs2115632680 |
80 | S>A | Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
| VAR_087211 | 172 | Q>del | NEDHISB [UniProt] | Yes | UniProt |
|
rs2115683966 RCV001824503 VAR_087212 |
173 | D>V | NEDHISB [UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
| VAR_087213 | 186 | E>del | NEDHISB; uncertain significance [UniProt] | Yes | UniProt |
|
rs1788864590 VAR_087214 RCV002246432 RCV001569146 |
224 | C>Y | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities NEDHISB [ClinVar, UniProt] | Yes |
ClinVar TOPMed dbSNP UniProt |
|
RCV002276505 rs1277446306 |
269 | N>K | GNAI1 associated Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar dbSNP gnomAD |
| VAR_087215 | 270 | K>N | NEDHISB [UniProt] | Yes | UniProt |
|
VAR_087216 rs2115712676 RCV002248391 |
270 | K>R | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities NEDHISB [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
RCV002248392 VAR_087217 rs2115712712 |
272 | D>G | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities NEDHISB [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
rs2115713066 RCV002273305 |
290 | Y>F | Neurodevelopmental disorder [ClinVar] | Yes |
ClinVar Ensembl dbSNP |
| VAR_087218 | 326 | A>P | NEDHISB [UniProt] | Yes | UniProt |
|
VAR_087219 RCV002248390 rs2115727431 |
332 | V>E | Neurodevelopmental disorder with hypotonia, impaired speech, and behavioral abnormalities NEDHISB; uncertain significance [ClinVar, UniProt] | Yes |
ClinVar Ensembl dbSNP UniProt |
|
rs1224009797 RCV001758457 |
5 | L>P | No |
ClinVar TOPMed dbSNP gnomAD |
|
| rs1162724911 | 6 | S>R | No |
TOPMed gnomAD |
|
| rs773988706 | 7 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1293940480 | 8 | E>A | No |
TOPMed gnomAD |
|
| rs1293940480 | 8 | E>V | No |
TOPMed gnomAD |
|
| rs760895601 | 9 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1397814226 | 9 | D>N | No | gnomAD | |
| COSM485597 | 10 | K>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1340678679 | 11 | A>E | No | gnomAD | |
| rs1340678679 | 11 | A>V | No | gnomAD | |
| rs1433259734 | 13 | V>L | No |
TOPMed gnomAD |
|
| rs776765120 | 15 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1787387719 | 17 | K>E | No | TOPMed | |
| COSM3640797 | 17 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs554748156 | 17 | K>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1787387941 | 21 | R>H | No | TOPMed | |
|
RCV001264620 rs1787388036 |
23 | L>missing | No |
ClinVar dbSNP |
|
| rs1787388145 | 25 | E>Q | No | TOPMed | |
| rs886804159 | 26 | D>A | No |
TOPMed gnomAD |
|
| rs1180244878 | 27 | G>S | No | gnomAD | |
| rs1417639782 | 33 | E>D | No | gnomAD | |
| rs1583996658 | 33 | E>K | No | Ensembl | |
| rs372783127 | 39 | L>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769148846 | 43 | E>K | No |
ExAC gnomAD |
|
| COSM453437 | 44 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1788434172 | 46 | K>* | No | Ensembl | |
| TCGA novel | 46 | K>M | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs770175485 | 46 | K>R | No |
ExAC gnomAD |
|
| rs1788434297 | 48 | T>A | No | gnomAD | |
|
rs1788434297 RCV001526691 |
48 | T>P | No |
ClinVar dbSNP gnomAD |
|
| TCGA novel | 49 | I>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1336698236 | 49 | I>V | No | TOPMed | |
| rs2115632430 | 55 | I>S | No | Ensembl | |
| rs1788436246 | 56 | I>M | No | Ensembl | |
| rs1788436294 | 57 | H>R | No | TOPMed | |
| rs1353589309 | 60 | G>A | No |
TOPMed gnomAD |
|
| rs1353589309 | 60 | G>D | No |
TOPMed gnomAD |
|
| rs779641348 | 62 | S>L | No |
ExAC gnomAD |
|
| rs1788436533 | 64 | E>K | No | TOPMed | |
| rs1340040890 | 65 | E>* | No | gnomAD | |
| rs751050803 | 65 | E>D | No |
ExAC gnomAD |
|
| TCGA novel | 67 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs550911078 | 67 | K>T | No | 1000Genomes | |
| TCGA novel | 70 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs367776716 | 71 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs367776716 | 71 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1319027288 | 74 | Y>C | No |
TOPMed gnomAD |
|
| rs773307363 | 77 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs771149857 | 77 | T>I | No |
ExAC gnomAD |
|
| rs771149857 | 77 | T>N | No |
ExAC gnomAD |
|
| COSM3929192 | 78 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1487731728 | 81 | I>V | No | gnomAD | |
| rs200973929 | 82 | I>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1193344371 | 82 | I>V | No | gnomAD | |
| rs1472132546 | 84 | I>V | No |
TOPMed gnomAD |
|
| rs182034954 | 87 | A>V | No | 1000Genomes | |
| rs1788437748 | 88 | M>L | No | gnomAD | |
| rs566542184 | 88 | M>T | No |
1000Genomes ExAC gnomAD |
|
|
rs1788437748 RCV002254045 |
88 | M>V | No |
ClinVar dbSNP gnomAD |
|
| rs1788437829 | 89 | G>R | No | gnomAD | |
| rs761858891 | 90 | R>K | No |
ExAC TOPMed gnomAD |
|
|
rs1400805039 COSM1673232 |
93 | I>T | large_intestine [Cosmic] | No |
cosmic curated gnomAD |
| rs955811912 | 93 | I>V | No |
TOPMed gnomAD |
|
| rs765148271 | 94 | D>A | No |
ExAC TOPMed gnomAD |
|
| rs765148271 | 94 | D>G | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 94 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1283536547 | 96 | G>D | No |
TOPMed gnomAD |
|
| rs1283536547 | 96 | G>V | No |
TOPMed gnomAD |
|
| rs367989878 | 97 | D>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs766033442 | 99 | A>D | No |
ExAC gnomAD |
|
| rs535549285 | 99 | A>T | No |
1000Genomes ExAC gnomAD |
|
| rs752261322 | 100 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754417098 | 100 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs548773475 | 101 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs548773475 COSM145453 |
101 | A>V | Variant assessed as Somatic; MODERATE impact. oesophagus haematopoietic_and_lymphoid_tissue [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1788636733 | 102 | D>G | No | Ensembl | |
| COSM6178773 | 102 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1258014515 | 103 | D>G | No | gnomAD | |
| rs1788636780 | 103 | D>N | No | Ensembl | |
| rs1258014515 | 103 | D>V | No | gnomAD | |
| rs1481220831 | 104 | A>T | No | gnomAD | |
| TCGA novel | 104 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs142908338 | 105 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
|
rs758691743 COSM269251 |
105 | R>H | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs1788637124 | 106 | Q>P | No | TOPMed | |
| TCGA novel | 109 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 111 | A>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs774459209 | 116 | E>D | No | Ensembl | |
| rs569973389 | 117 | G>A | No | 1000Genomes | |
| rs1476883165 | 119 | M>K | No | gnomAD | |
| rs747063794 | 119 | M>V | No |
ExAC gnomAD |
|
| rs986358825 | 120 | T>I | No | TOPMed | |
| rs1170003605 | 121 | A>T | No | gnomAD | |
| COSM6178772 | 122 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1788637809 | 123 | L>F | No | Ensembl | |
| rs1788637860 | 123 | L>P | No | Ensembl | |
| rs2115668506 | 124 | A>P | No | Ensembl | |
| rs773134896 | 124 | A>V | No |
ExAC gnomAD |
|
| rs1584045095 | 125 | G>R | No | TOPMed | |
| rs1788638071 | 126 | V>I | No | TOPMed | |
| rs770957575 | 127 | I>M | No |
ExAC gnomAD |
|
| rs35545354 | 127 | I>T | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1335604046 | 127 | I>V | No | gnomAD | |
| rs1788638378 | 128 | K>R | No | Ensembl | |
| rs1174216714 | 130 | L>S | No | TOPMed | |
| rs1584045143 | 131 | W>* | No | Ensembl | |
| rs1584045141 | 131 | W>G | No | Ensembl | |
| rs1397620498 | 132 | K>E | No | gnomAD | |
| rs942163781 | 132 | K>R | No | TOPMed | |
| rs1722838527 | 133 | D>G | No | TOPMed | |
| rs759363974 | 133 | D>N | No |
ExAC gnomAD |
|
| rs771688081 | 135 | G>S | No |
ExAC gnomAD |
|
| rs775142147 | 136 | V>I | No |
ExAC gnomAD |
|
| rs760188122 | 137 | Q>K | No |
ExAC gnomAD |
|
| rs763521326 | 138 | A>S | No |
ExAC gnomAD |
|
| rs763521326 | 138 | A>T | No |
ExAC gnomAD |
|
| rs1271356769 | 139 | C>S | No | gnomAD | |
| rs191814694 | 140 | F>L | No |
1000Genomes TOPMed gnomAD |
|
| rs145021139 | 141 | N>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs145021139 | 141 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1788639490 | 142 | R>I | No | Ensembl | |
| rs758884255 | 144 | R>Q | No |
ExAC gnomAD |
|
| TCGA novel | 145 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1584045219 | 147 | Q>H | No | Ensembl | |
| rs751761285 | 151 | S>C | No |
ExAC gnomAD |
|
| rs368834370 | 152 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1314128142 | 153 | A>V | No | Ensembl | |
| rs756206752 | 158 | D>E | No |
ExAC gnomAD |
|
| rs2115683838 | 160 | D>A | No | Ensembl | |
| rs778777547 | 160 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1788727649 | 161 | R>K | No |
TOPMed gnomAD |
|
| rs1407827636 | 162 | I>V | No | TOPMed | |
| rs745806214 | 165 | P>A | No |
ExAC TOPMed gnomAD |
|
| rs745806214 | 165 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs897276806 | 166 | N>D | No | Ensembl | |
| rs1788727810 | 167 | Y>D | No | TOPMed | |
| rs1387120752 | 169 | P>L | No |
TOPMed gnomAD |
|
|
COSM3923998 rs1204209552 |
169 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1420173226 | 171 | Q>E | No | TOPMed | |
| rs1180471133 | 174 | V>I | No | TOPMed | |
| rs1408290484 | 177 | T>I | No | gnomAD | |
| rs1788728185 | 178 | R>T | No | Ensembl | |
| rs1562842351 | 181 | T>I | No | Ensembl | |
| rs1788728360 | 182 | T>I | No | Ensembl | |
|
rs2115684055 RCV001765393 |
183 | G>missing | No |
ClinVar dbSNP |
|
| COSM4911810 | 184 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1173391823 | 185 | V>I | No | gnomAD | |
| rs1584048872 | 188 | H>R | No | Ensembl | |
| TCGA novel | 190 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 190 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1243909467 | 194 | L>R | No |
TOPMed gnomAD |
|
| COSM4848672 | 201 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1337409948 | 204 | Q>E | No | gnomAD | |
| rs1788863320 | 204 | Q>R | No | Ensembl | |
| COSM3431776 | 205 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs910988907 COSM196932 |
208 | R>Q | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl NCI-TCGA |
| COSM1092027 | 213 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1562844525 | 216 | E>K | No | Ensembl | |
| rs17852392 | 219 | T>A | No | Ensembl | |
| rs772844909 | 219 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 220 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM196933 rs1211791957 |
220 | A>V | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA TOPMed gnomAD |
| rs549996015 | 223 | F>I | No |
1000Genomes ExAC gnomAD |
|
| rs774942165 | 225 | V>A | No | ExAC | |
| rs1025287036 | 225 | V>I | No | TOPMed | |
| rs1187632806 | 228 | S>G | No |
TOPMed gnomAD |
|
|
COSM3394899 rs1403981151 |
231 | D>N | pancreas [Cosmic] | No |
cosmic curated gnomAD |
| rs922293765 | 234 | L>V | No | TOPMed | |
| rs764370373 | 235 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs2115707096 | 238 | E>D | No | Ensembl | |
| rs753886455 | 240 | M>T | No |
ExAC gnomAD |
|
| rs1788866072 | 240 | M>V | No | gnomAD | |
|
rs1788895532 COSM1092029 |
242 | R>* | endometrium [Cosmic] | No |
cosmic curated gnomAD |
| rs1788895532 | 242 | R>G | No | gnomAD | |
| rs911562127 | 247 | M>K | No | TOPMed | |
| rs1788895878 | 251 | D>E | No |
TOPMed gnomAD |
|
| rs1788895823 | 251 | D>G | No |
TOPMed gnomAD |
|
| rs1331183452 | 253 | I>M | No | TOPMed | |
| rs1788895971 | 254 | C>Y | No | TOPMed | |
| rs1466308269 | 256 | N>D | No |
TOPMed gnomAD |
|
| rs1163446805 | 256 | N>S | No | gnomAD | |
| TCGA novel | 257 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4420716 | 257 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1346935768 | 259 | F>V | No | gnomAD | |
| rs12721453 | 261 | D>N | No | Ensembl | |
| rs775831477 | 262 | T>K | No |
ExAC gnomAD |
|
| COSM4876466 | 264 | I>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 265 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 266 | L>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761843186 | 271 | K>N | No |
ExAC gnomAD |
|
|
RCV001816465 rs2115712706 |
272 | D>N | No |
ClinVar Ensembl dbSNP |
|
| rs975250753 | 273 | L>V | No |
TOPMed gnomAD |
|
| rs535650213 | 274 | F>I | No |
1000Genomes ExAC gnomAD |
|
| rs923267671 | 276 | E>* | No |
TOPMed gnomAD |
|
| rs954904149 | 276 | E>G | No |
TOPMed gnomAD |
|
|
rs2115712787 RCV001774462 |
276 | E>L | No |
ClinVar Ensembl dbSNP |
|
| rs954904149 | 276 | E>V | No |
TOPMed gnomAD |
|
|
rs1244226395 RCV000598656 CA575882515 |
278 | I>missing | No |
ClinGen ClinVar dbSNP |
|
| rs1180055744 | 278 | I>T | No | gnomAD | |
| rs751395120 | 278 | I>V | No |
ExAC gnomAD |
|
| rs1363490208 | 279 | K>E | No | gnomAD | |
| rs1470617220 | 279 | K>R | No |
TOPMed gnomAD |
|
| rs1788897709 | 280 | K>N | No |
TOPMed gnomAD |
|
| rs1788897660 | 280 | K>R | No | Ensembl | |
| TCGA novel | 282 | P>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs908500528 | 282 | P>L | No | TOPMed | |
| rs908500528 | 282 | P>R | No | TOPMed | |
| TCGA novel | 283 | L>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs755993821 COSM1092030 |
284 | T>I | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA gnomAD |
| rs1788898088 | 285 | I>M | No | Ensembl | |
| rs777524714 | 285 | I>V | No |
ExAC gnomAD |
|
| rs1788898155 | 286 | C>Y | No | gnomAD | |
| rs367981356 | 287 | Y>C | No |
ExAC gnomAD |
|
| rs1454400629 | 287 | Y>H | No | gnomAD | |
| rs772836802 | 288 | P>A | No |
TOPMed gnomAD |
|
| rs17852394 | 288 | P>Q | No | Ensembl | |
| rs772836802 | 288 | P>S | No |
TOPMed gnomAD |
|
| rs772836802 | 288 | P>T | No |
TOPMed gnomAD |
|
| TCGA novel | 289 | E>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1334841117 | 291 | A>T | No | gnomAD | |
| rs767511386 | 292 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs767511386 | 292 | G>E | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 293 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1788990033 | 295 | T>I | No | TOPMed | |
| TCGA novel | 295 | T>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1227100605 | 296 | Y>C | No |
TOPMed gnomAD |
|
| rs753742190 | 296 | Y>H | No |
ExAC gnomAD |
|
| rs1434894043 | 297 | E>D | No |
TOPMed gnomAD |
|
| rs764841177 | 299 | A>T | No |
ExAC gnomAD |
|
|
COSM748032 rs1788990508 |
302 | Y>C | lung Variant assessed as Somatic; MODERATE impact. [Cosmic, NCI-TCGA] | No |
NCI-TCGA Cosmic cosmic curated TOPMed gnomAD |
| TCGA novel | 304 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 309 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM748031 | 310 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372256817 | 311 | N>D | No |
ESP TOPMed gnomAD |
|
| rs1584059315 | 311 | N>S | No | Ensembl | |
| rs1788990833 | 314 | K>T | No | Ensembl | |
|
rs1788990885 COSM1313313 |
315 | D>N | Variant assessed as Somatic; MODERATE impact. urinary_tract [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated TOPMed |
| rs1369121503 | 319 | I>T | No | gnomAD | |
| TCGA novel | 320 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1396776828 | 320 | Y>H | No | TOPMed | |
| rs1788991352 | 321 | T>A | No |
TOPMed gnomAD |
|
| rs1384388891 | 321 | T>I | No |
TOPMed gnomAD |
|
| rs1788991452 | 322 | H>Y | No | TOPMed | |
| rs755277179 | 325 | C>R | No |
ExAC gnomAD |
|
| COSM1092031 | 326 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1390724642 | 327 | T>A | No |
TOPMed gnomAD |
|
| COSM748030 | 331 | N>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs781525050 | 332 | V>M | No |
ExAC gnomAD |
|
| rs772886652 | 333 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs552102809 | 335 | V>G | No | 1000Genomes | |
| rs770033700 | 336 | F>L | No |
ExAC gnomAD |
|
| COSM4687382 | 336 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1294148085 | 338 | A>V | No | gnomAD | |
| rs773328998 | 339 | V>L | No |
ExAC gnomAD |
|
| rs771135868 | 343 | I>V | No |
ExAC gnomAD |
|
| rs1383142864 | 344 | I>V | No | gnomAD | |
| rs1788993053 | 346 | N>D | No | Ensembl | |
| rs1788993100 | 346 | N>K | No | Ensembl | |
| TCGA novel | 346 | N>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1189213040 | 348 | L>V | No | Ensembl | |
| rs1788994023 | 350 | D>Y | No | Ensembl | |
| COSM3833293 | 351 | C>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM339898 | 352 | G>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 352 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 354 | F>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 354 | F>L | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1092032 | 354 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1351222111 | 354 | F>S | No | gnomAD | |
| rs1451026867 | 355 | F>Q | No | gnomAD | |
| rs760674936 | 355 | F>Y | No |
ExAC gnomAD |
No associated diseases with P63096
Functions
10 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell cortex | The region of a cell that lies just beneath the plasma membrane and often, but not always, contains a network of actin filaments and associated proteins. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| heterotrimeric G-protein complex | Any of a family of heterotrimeric GTP-binding and hydrolyzing proteins; they belong to a superfamily of GTPases that includes monomeric proteins such as EF-Tu and RAS. Heterotrimeric G-proteins consist of three subunits; the alpha subunit contains the guanine nucleotide binding site and possesses GTPase activity; the beta and gamma subunits are tightly associated and function as a beta-gamma heterodimer; extrinsic plasma membrane proteins (cytoplasmic face) that function as a complex to transduce signals from G protein-coupled receptors to an effector protein. |
| lysosomal membrane | The lipid bilayer surrounding the lysosome and separating its contents from the cell cytoplasm. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| D2 dopamine receptor binding | Binding to a D2 dopamine receptor. |
| G protein-coupled receptor binding | Binding to a G protein-coupled receptor. |
| G protein-coupled serotonin receptor binding | Binding to a metabotropic serotonin receptor. |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the inhibition of adenylyl cyclase activity and a subsequent decrease in the intracellular concentration of cyclic AMP (cAMP). |
| adenylate cyclase-modulating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation or inhibition of adenylyl cyclase activity and a subsequent change in the intracellular concentration of cyclic AMP (cAMP). |
| cell cycle | The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| cellular response to forskolin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a forskolin stimulus. |
| G protein-coupled receptor signaling pathway | The series of molecular signals initiated by a ligand binding to its receptor, in which the activated receptor promotes the exchange of GDP for GTP on the alpha-subunit of an associated heterotrimeric G-protein complex. The GTP-bound activated alpha-G-protein then dissociates from the beta- and gamma-subunits to further transmit the signal within the cell. The pathway begins with receptor-ligand interaction, and ends with regulation of a downstream cellular process. The pathway can start from the plasma membrane, Golgi or nuclear membrane. |
| positive regulation of protein localization to cell cortex | Any process that activates or increases the frequency, rate or extent of protein localization to cell cortex. |
| regulation of cAMP-mediated signaling | Any process which modulates the frequency, rate or extent of cAMP-mediated signaling. |
| regulation of mitotic spindle organization | Any process that modulates the rate, frequency or extent of the assembly, arrangement of constituent parts, or disassembly of the microtubule spindle during a mitotic cell cycle. |
| response to peptide hormone | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a peptide hormone stimulus. A peptide hormone is any of a class of peptides that are secreted into the blood stream and have endocrine functions in living animals. |
62 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P0C7Q4 | GNAT3 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04696 | GNAT2 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P04695 | GNAT1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P08239 | GNAO1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P63097 | GNAI1 | Guanine nucleotide-binding protein G | Bos taurus (Bovine) | SS |
| P50147 | GNAI2 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P50146 | GNAI1 | Guanine nucleotide-binding protein G | Gallus gallus (Chicken) | SS |
| P16378 | Galphao | G protein alpha o subunit | Drosophila melanogaster (Fruit fly) | SS |
| P25157 | cta | Guanine nucleotide-binding protein subunit alpha homolog | Drosophila melanogaster (Fruit fly) | SS |
| P20353 | Galphai | G protein alpha i subunit | Drosophila melanogaster (Fruit fly) | SS |
| Q14344 | GNA13 | Guanine nucleotide-binding protein subunit alpha-13 | Homo sapiens (Human) | SS |
| Q03113 | GNA12 | Guanine nucleotide-binding protein subunit alpha-12 | Homo sapiens (Human) | SS |
| P29992 | GNA11 | Guanine nucleotide-binding protein subunit alpha-11 | Homo sapiens (Human) | SS |
| P30679 | GNA15 | Guanine nucleotide-binding protein subunit alpha-15 | Homo sapiens (Human) | SS |
| O95837 | GNA14 | Guanine nucleotide-binding protein subunit alpha-14 | Homo sapiens (Human) | SS |
| P50148 | GNAQ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P38405 | GNAL | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| A8MTJ3 | GNAT3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19087 | GNAT2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P11488 | GNAT1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P04899 | GNAI2 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P08754 | GNAI3 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P19086 | GNAZ | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| P09471 | GNAO1 | Guanine nucleotide-binding protein G | Homo sapiens (Human) | SS |
| Q9DC51 | Gnai3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27600 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Mus musculus (Mouse) | SS |
| P20612 | Gnat1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P27601 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Mus musculus (Mouse) | SS |
| Q3V3I2 | Gnat3 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| O70443 | Gnaz | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P50149 | Gnat2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P08752 | Gnai2 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P18872 | Gnao1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| B2RSH2 | Gnai1 | Guanine nucleotide-binding protein G | Mus musculus (Mouse) | SS |
| P93564 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum tuberosum (Potato) | SS |
| Q63210 | Gna12 | Guanine nucleotide-binding protein subunit alpha-12 | Rattus norvegicus (Rat) | SS |
| P19627 | Gnaz | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P29348 | Gnat3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P04897 | Gnai2 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P59215 | Gnao1 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| P08753 | Gnai3 | Guanine nucleotide-binding protein G | Rattus norvegicus (Rat) | SS |
| Q6Q7Y5 | Gna13 | Guanine nucleotide-binding protein subunit alpha-13 | Rattus norvegicus (Rat) | SS |
| P10824 | Gnai1 | Guanine nucleotide-binding protein G subunit alpha-1 | Rattus norvegicus (Rat) | EV |
| Q0DJ33 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Oryza sativa subsp. japonica (Rice) | SS |
| O76584 | gpa-11 | Guanine nucleotide-binding protein alpha-11 subunit | Caenorhabditis elegans | SS |
| P51875 | goa-1 | Guanine nucleotide-binding protein G | Caenorhabditis elegans | SS |
| Q18434 | odr-3 | Guanine nucleotide-binding protein alpha-17 subunit | Caenorhabditis elegans | SS |
| Q9BIG4 | gpa-10 | Guanine nucleotide-binding protein alpha-10 subunit | Caenorhabditis elegans | PR |
| P28052 | gpa-3 | Guanine nucleotide-binding protein alpha-3 subunit | Caenorhabditis elegans | SS |
| Q9XTB2 | gpa-13 | Guanine nucleotide-binding protein alpha-13 subunit | Caenorhabditis elegans | SS |
| Q93743 | gpa-6 | Guanine nucleotide-binding protein alpha-6 subunit | Caenorhabditis elegans | SS |
| Q20907 | gpa-8 | Guanine nucleotide-binding protein alpha-8 subunit | Caenorhabditis elegans | PR |
| P91907 | gpa-15 | Guanine nucleotide-binding protein alpha-15 subunit | Caenorhabditis elegans | SS |
| Q21917 | gpa-7 | Guanine nucleotide-binding protein alpha-7 subunit | Caenorhabditis elegans | SS |
| P28051 | gpa-1 | Guanine nucleotide-binding protein alpha-1 subunit | Caenorhabditis elegans | SS |
| Q9BIG5 | gpa-4 | Guanine nucleotide-binding protein alpha-4 subunit | Caenorhabditis elegans | SS |
| Q9N2V6 | gpa-16 | Guanine nucleotide-binding protein alpha-16 subunit | Caenorhabditis elegans | SS |
| P93163 | GPA2 | Guanine nucleotide-binding protein alpha-2 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| P49084 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Glycine max (Soybean) (Glycine hispida) | SS |
| O80462 | XLG1 | Extra-large guanine nucleotide-binding protein 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P18064 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P26981 | GPA1 | Guanine nucleotide-binding protein alpha-1 subunit | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGCTLSAEDK | AAVERSKMID | RNLREDGEKA | AREVKLLLLG | AGESGKSTIV | KQMKIIHEAG |
| 70 | 80 | 90 | 100 | 110 | 120 |
| YSEEECKQYK | AVVYSNTIQS | IIAIIRAMGR | LKIDFGDSAR | ADDARQLFVL | AGAAEEGFMT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| AELAGVIKRL | WKDSGVQACF | NRSREYQLND | SAAYYLNDLD | RIAQPNYIPT | QQDVLRTRVK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TTGIVETHFT | FKDLHFKMFD | VGGQRSERKK | WIHCFEGVTA | IIFCVALSDY | DLVLAEDEEM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| NRMHESMKLF | DSICNNKWFT | DTSIILFLNK | KDLFEEKIKK | SPLTICYPEY | AGSNTYEEAA |
| 310 | 320 | 330 | 340 | 350 | |
| AYIQCQFEDL | NKRKDTKEIY | THFTCATDTK | NVQFVFDAVT | DVIIKNNLKD | CGLF |