P62071
Gene name |
Rras2 |
Protein name |
Ras-related protein R-Ras2 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:66922 |
EC number |
|
Protein Class |
|
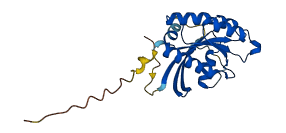
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for P62071
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-P62071-F1 | Predicted | AlphaFoldDB |
11 variants for P62071
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388944007 | 40 | V>E | No | EVA | |
| rs3398180535 | 47 | I>M | No | EVA | |
| rs3388942366 | 58 | D>N | No | EVA | |
| rs3388928359 | 72 | Q>E | No | EVA | |
| rs3388943030 | 92 | V>I | No | EVA | |
| rs3388943001 | 106 | K>Q | No | EVA | |
| rs3388928365 | 115 | K>* | No | EVA | |
| rs3388913167 | 148 | Q>R | No | EVA | |
| rs3388935461 | 161 | R>K | No | EVA | |
| rs3398412696 | 179 | Q>H | No | EVA | |
| rs3388939146 | 190 | T>I | No | EVA |
No associated diseases with P62071
1 regional properties for P62071
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 14 - 170 | IPR005225 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| cell migration | The controlled self-propelled movement of a cell from one site to a destination guided by molecular cues. Cell migration is a central process in the development and maintenance of multicellular organisms. |
| positive regulation of Schwann cell migration | Any process that activates or increases the frequency, rate or extent of Schwann cell migration. |
| Ras protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Ras superfamily of proteins switching to a GTP-bound active state. |
| regulation of neuron death | Any process that modulates the frequency, rate or extent of neuron death. |
| Schwann cell migration | The orderly movement of a Schwann cell from one site to another. A Schwann cell is a glial cell that ensheathes axons of neuron in the peripheral nervous system and is necessary for their maintainance and function. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| P62070 | RRAS2 | Ras-related protein R-Ras2 | Homo sapiens (Human) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAAGWRDGS | GQEKYRLVVV | GGGGVGKSAL | TIQFIQSYFV | TDYDPTIEDS | YTKQCVIDDR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AARLDILDTA | GQEEFGAMRE | QYMRTGEGFL | LVFSVTDRGS | FEEIYKFQRQ | ILRVKDRDEF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PMILIGNKAD | LDHQRQVTQE | EGQQLARQLK | VTYMEASAKI | RMNVDQAFHE | LVRVIRKFQE |
| 190 | 200 | ||||
| QECPPSPEPT | RKEKDKKGCH | CVIF |