P62070
Gene name |
RRAS2 |
Protein name |
Ras-related protein R-Ras2 |
Names |
Ras-like protein TC21, Teratocarcinoma oncogene |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:22800 |
EC number |
|
Protein Class |
|
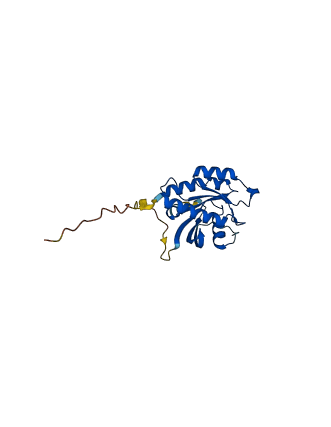
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for P62070
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2ERY | X-ray | 170 A | A/B | 12-181 | PDB |
| AF-P62070-F1 | Predicted | AlphaFoldDB |
122 variants for P62070
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs1591495776 RCV000852395 |
22 | G>missing | Noonan syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
CA379862795 RCV000852396 rs1591495779 VAR_083149 |
23 | G>V | Noonan syndrome NS12; increased MAPK signaling [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000852399 RCV001265738 rs1591495767 RCV003117554 RCV000853184 |
24 | G>missing | Noonan syndrome Noonan syndrome 12 Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
| VAR_083150 | 24 | G>GGGG | NS12; increased MAPK signaling [UniProt] | Yes | UniProt |
| VAR_083151 | 26 | G>GGVG | NS12; increased MAPK signaling; results in craniofacial patterning defects when expressed in zebrafish [UniProt] | Yes | UniProt |
|
rs782457908 RCV000853183 COSM3719717 CA5894328 RCV000852397 VAR_083152 |
70 | A>T | Variant assessed as Somatic; 0.0 impact. Noonan syndrome Noonan syndrome 12 haematopoietic_and_lymphoid_tissue NS12; decreased GAP-stimulated GTPase activity leading to an accumulation of RRAS2 in its GTP-bound active state; increased MAPK signaling; loss of interaction with RASSF5 [NCI-TCGA, ClinVar, Cosmic, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt ExAC NCI-TCGA dbSNP gnomAD |
| VAR_083153 | 72 | Q>H | NS12; associated in cis with C-75; increased MAPK signaling; results in craniofacial patterning defects when expressed in zebrafish; results in craniofacial patterning defects in zebrafish when associated with C-75 [UniProt] | Yes | UniProt |
|
CA120437 COSM687135 rs113954997 VAR_006848 RCV000852398 RCV000010054 RCV001072115 |
72 | Q>L | lung Noonan syndrome Noonan syndrome 12 Variant assessed as Somatic; impact. endometrium Neoplasm of ovary NS12; also found as somatic mutation in ovarian cancer; increased MAPK signaling; results in craniofacial patterning defects when expressed in zebrafish [Cosmic, ClinVar, NCI-TCGA, UniProt] | Yes |
ClinGen cosmic curated ClinVar UniProt Ensembl NCI-TCGA dbSNP |
| TCGA novel | 3 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782151181 CA5894417 |
5 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1439228169 CA379862911 |
7 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA5894416 rs781902160 |
10 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA379862882 rs1174339583 |
11 | G>S | No |
ClinGen TOPMed |
|
|
CA379862830 rs1554955854 |
18 | V>L | No |
ClinGen gnomAD |
|
|
CA379862819 rs1554955852 |
19 | V>G | No |
ClinGen Ensembl |
|
|
CA379862824 rs1554955853 |
19 | V>M | No |
ClinGen gnomAD |
|
|
CA379862814 rs1554955851 |
20 | V>G | No |
ClinGen Ensembl |
|
|
rs1554955849 CA379862805 |
22 | G>R | No |
ClinGen gnomAD |
|
|
rs1554955845 CA379862780 |
26 | G>S | No |
ClinGen gnomAD |
|
|
rs1591495759 CA379862749 |
31 | T>S | No |
ClinGen Ensembl |
|
|
CA379862738 rs1396949099 |
32 | I>M | No |
ClinGen TOPMed |
|
|
rs1554955843 CA379862732 |
33 | Q>L | No |
ClinGen gnomAD |
|
|
rs1448641952 CA379862721 |
35 | I>L | No |
ClinGen TOPMed |
|
|
CA379862717 rs1328938728 |
35 | I>T | No |
ClinGen TOPMed |
|
|
CA5894356 rs782273898 |
40 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs782670869 CA5894355 |
41 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 44 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs913808099 CA218175411 |
45 | P>L | No |
ClinGen Ensembl |
|
|
rs1372841871 CA379860882 |
48 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 56 | V>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA379860819 rs1554946506 |
57 | I>V | No |
ClinGen gnomAD |
|
|
CA5894354 rs552513136 |
60 | R>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 60 | R>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 62 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs782597266 CA5894352 |
63 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782597266 CA379860777 |
63 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs782189872 CA5894353 |
63 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs782481088 CA5894351 |
64 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA379860670 rs1160017680 |
71 | G>E | No |
ClinGen TOPMed |
|
| TCGA novel | 72 | Q>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs113954997 CA218175344 |
72 | Q>R | No |
ClinGen Ensembl |
|
| TCGA novel | 74 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| VAR_083154 | 75 | F>C | likely benign variant; associated in cis with H-72 in a patient with Noonan syndrome; has no effect on MAPK signaling; has no effect on craniofacial patterning when expressed in zebrafish; results in craniofacial patterning defects in zebrafish when associated with H-72 [UniProt] | No | UniProt |
|
CA218175342 rs1009231474 |
75 | F>L | No |
ClinGen Ensembl |
|
|
CA218175341 rs202147465 |
77 | A>G | No |
ClinGen Ensembl |
|
|
rs1554946344 CA379860571 |
78 | M>L | No |
ClinGen gnomAD |
|
|
rs782611291 CA5894326 |
85 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA379860446 rs1554946338 |
86 | G>D | No |
ClinGen gnomAD |
|
|
rs1554946335 CA379860441 |
87 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs141697442 CA5894324 |
88 | G>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1554946329 CA379860391 |
91 | L>M | No |
ClinGen gnomAD |
|
|
rs782166268 CA5894322 |
92 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 93 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 94 | S>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA379860337 rs1306675531 |
95 | V>I | No |
ClinGen TOPMed |
|
|
rs1352874625 CA379860309 |
97 | D>G | No |
ClinGen TOPMed |
|
|
rs1554946327 CA379860316 |
97 | D>N | No |
ClinGen gnomAD |
|
| TCGA novel | 98 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA218175338 rs944526520 |
99 | G>R | No |
ClinGen gnomAD |
|
|
rs1554946299 CA379860231 |
102 | E>Q | No |
ClinGen gnomAD |
|
|
rs1246077414 CA379860222 |
103 | E>G | No |
ClinGen TOPMed |
|
|
CA5894300 rs782567491 |
103 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs571234555 CA5894299 |
105 | Y>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA379860201 rs1554946294 |
106 | K>* | No |
ClinGen gnomAD |
|
|
rs781814822 CA5894298 |
110 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
rs1554946289 CA379860130 |
115 | K>R | No |
ClinGen gnomAD |
|
|
rs782490571 CA5894296 COSM181392 |
117 | R>C | Variant assessed as Somatic; 4.657e-05 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs782490571 CA5894297 |
117 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs566703225 CA5894295 |
117 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs566703225 CA379860117 |
117 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1554946283 CA379860082 |
122 | M>V | No |
ClinGen gnomAD |
|
|
rs782790768 CA5894294 |
123 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA379860063 rs782171884 |
124 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs370045365 CA218175327 |
125 | I>V | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs781928921 CA5894292 |
134 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA5894275 rs549966258 |
138 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1015087676 CA217901375 |
142 | G>R | No |
ClinGen gnomAD |
|
|
rs782004900 CA5894272 |
145 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs377321245 COSM925277 CA5894269 |
147 | R>Q | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA5894270 rs782047286 |
147 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5894268 rs138886317 |
148 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs782092871 CA5894267 |
150 | K>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA379733017 rs1554944335 |
151 | V>I | No |
ClinGen gnomAD |
|
|
CA5894264 rs782269601 |
154 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA379732964 rs1554944332 |
154 | M>V | No |
ClinGen gnomAD |
|
|
CA5894263 rs782694684 |
156 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA379732905 rs1213773767 |
158 | A>G | No |
ClinGen TOPMed |
|
|
rs1316363049 CA379732915 COSM1352819 |
158 | A>T | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA TOPMed gnomAD |
|
rs1554944326 CA379732838 |
162 | M>R | No |
ClinGen gnomAD |
|
|
CA5894262 rs782311837 |
162 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs372963005 CA379732815 |
164 | V>I | No |
ClinGen ESP ExAC gnomAD |
|
|
rs372963005 CA5894261 |
164 | V>L | No |
ClinGen ESP ExAC gnomAD |
|
|
rs781860392 CA5894258 |
169 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs1465550850 CA379732710 |
170 | E>Q | No |
ClinGen TOPMed |
|
|
rs782654813 CA5894257 |
171 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA5894255 rs781787510 |
172 | V>F | No |
ClinGen ExAC |
|
|
CA5894253 rs782079709 |
173 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA5894254 rs782726211 |
173 | R>W | Variant assessed as Somatic; 4.66e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1241753346 CA379732639 |
175 | I>V | No |
ClinGen TOPMed |
|
|
rs782754561 CA5894251 |
176 | R>T | No |
ClinGen ExAC gnomAD |
|
|
CA379732040 rs1554943977 |
177 | K>E | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 178 | F>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1386909460 CA379731995 |
180 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1554943974 CA379731980 |
181 | Q>E | No |
ClinGen gnomAD |
|
|
CA379731931 rs1384357948 |
184 | P>R | No |
ClinGen TOPMed |
|
|
CA379731925 rs1564950642 |
185 | P>S | No |
ClinGen Ensembl |
|
|
rs1446700718 CA379731902 |
187 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1446700718 CA379731899 |
187 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1446700718 CA379731900 |
187 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA5894227 rs781970568 |
191 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA5894228 rs370564233 |
191 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 193 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA5894225 rs782141522 |
193 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA5894226 rs782761955 |
193 | E>K | No |
ClinGen ExAC |
|
|
rs1554943963 CA379731819 |
194 | K>E | No |
ClinGen gnomAD |
|
|
CA379731813 rs1233671138 |
194 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA5894224 rs569248551 |
195 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 195 | D>Y | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1554943958 CA379731776 |
197 | K>E | No |
ClinGen gnomAD |
|
|
rs1554943957 CA379731770 |
197 | K>T | No |
ClinGen gnomAD |
|
|
rs1564950576 CA379731745 |
199 | C>Y | No |
ClinGen Ensembl |
|
|
CA5894223 rs782423525 |
200 | H>Y | No |
ClinGen ExAC gnomAD |
2 associated diseases with P62070
[MIM: 167000]: Ovarian cancer (OC)
The term ovarian cancer defines malignancies originating from ovarian tissue. Although many histologic types of ovarian tumors have been described, epithelial ovarian carcinoma is the most common form. Ovarian cancers are often asymptomatic and the recognized signs and symptoms, even of late-stage disease, are vague. Consequently, most patients are diagnosed with advanced disease. {ECO:0000269|PubMed:8052619}. Note=Disease susceptibility is associated with variants affecting the gene represented in this entry.
[MIM: 618624]: Noonan syndrome 12 (NS12)
A form of Noonan syndrome, a disease characterized by short stature, facial dysmorphic features such as hypertelorism, a downward eyeslant and low-set posteriorly rotated ears, and a high incidence of congenital heart defects and hypertrophic cardiomyopathy. Other features can include a short neck with webbing or redundancy of skin, deafness, motor delay, variable intellectual deficits, multiple skeletal defects, cryptorchidism, and bleeding diathesis. Individuals with Noonan syndrome are at risk of juvenile myelomonocytic leukemia, a myeloproliferative disorder characterized by excessive production of myelomonocytic cells. NS12 inheritance is autosomal dominant. There is considerable variability in severity. {ECO:0000269|PubMed:31130282, ECO:0000269|PubMed:31130285}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- The term ovarian cancer defines malignancies originating from ovarian tissue. Although many histologic types of ovarian tumors have been described, epithelial ovarian carcinoma is the most common form. Ovarian cancers are often asymptomatic and the recognized signs and symptoms, even of late-stage disease, are vague. Consequently, most patients are diagnosed with advanced disease. {ECO:0000269|PubMed:8052619}. Note=Disease susceptibility is associated with variants affecting the gene represented in this entry.
- A form of Noonan syndrome, a disease characterized by short stature, facial dysmorphic features such as hypertelorism, a downward eyeslant and low-set posteriorly rotated ears, and a high incidence of congenital heart defects and hypertrophic cardiomyopathy. Other features can include a short neck with webbing or redundancy of skin, deafness, motor delay, variable intellectual deficits, multiple skeletal defects, cryptorchidism, and bleeding diathesis. Individuals with Noonan syndrome are at risk of juvenile myelomonocytic leukemia, a myeloproliferative disorder characterized by excessive production of myelomonocytic cells. NS12 inheritance is autosomal dominant. There is considerable variability in severity. {ECO:0000269|PubMed:31130282, ECO:0000269|PubMed:31130285}. Note=The disease is caused by variants affecting the gene represented in this entry.
1 regional properties for P62070
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 14 - 170 | IPR005225 |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| GDP binding | Binding to GDP, guanosine 5'-diphosphate. |
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| osteoblast differentiation | The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, a mesodermal or neural crest cell that gives rise to bone. |
| positive regulation of Schwann cell migration | Any process that activates or increases the frequency, rate or extent of Schwann cell migration. |
| Ras protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Ras superfamily of proteins switching to a GTP-bound active state. |
| regulation of neuron death | Any process that modulates the frequency, rate or extent of neuron death. |
| Schwann cell migration | The orderly movement of a Schwann cell from one site to another. A Schwann cell is a glial cell that ensheathes axons of neuron in the peripheral nervous system and is necessary for their maintainance and function. |
34 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P08642 | HRAS | GTPase HRas | Gallus gallus (Chicken) | SS |
| Q6T310 | RASL11A | Ras-like protein family member 11A | Homo sapiens (Human) | PR |
| Q8IYK8 | REM2 | GTP-binding protein REM 2 | Homo sapiens (Human) | PR |
| P55040 | GEM | GTP-binding protein GEM | Homo sapiens (Human) | PR |
| Q6IQ22 | RAB12 | Ras-related protein Rab-12 | Homo sapiens (Human) | PR |
| Q9BU20 | CPLANE2 | Ciliogenesis and planar polarity effector 2 | Homo sapiens (Human) | PR |
| Q96HU8 | DIRAS2 | GTP-binding protein Di-Ras2 | Homo sapiens (Human) | PR |
| P11234 | RALB | Ras-related protein Ral-B | Homo sapiens (Human) | PR |
| P11233 | RALA | Ras-related protein Ral-A | Homo sapiens (Human) | PR |
| Q99578 | RIT2 | GTP-binding protein Rit2 | Homo sapiens (Human) | PR |
| P01116 | KRAS | GTPase KRas | Homo sapiens (Human) | EV |
| P01112 | HRAS | GTPase HRas | Homo sapiens (Human) | SS |
| Q9JIW9 | Ralb | Ras-related protein Ral-B | Mus musculus (Mouse) | PR |
| Q61411 | Hras | GTPase HRas | Mus musculus (Mouse) | SS |
| P32883 | Kras | GTPase KRas | Mus musculus (Mouse) | SS |
| O08989 | Mras | Ras-related protein M-Ras | Mus musculus (Mouse) | PR |
| Q5PR73 | Diras2 | GTP-binding protein Di-Ras2 | Mus musculus (Mouse) | PR |
| Q91Z61 | Diras1 | GTP-binding protein Di-Ras1 | Mus musculus (Mouse) | PR |
| P35283 | Rab12 | Ras-related protein Rab-12 | Mus musculus (Mouse) | PR |
| Q08AT1 | Rasl12 | Ras-like protein family member 12 | Mus musculus (Mouse) | PR |
| A2A825 | Cplane2 | Ciliogenesis and planar polarity effector 2 | Mus musculus (Mouse) | PR |
| P55041 | Gem | GTP-binding protein GEM | Mus musculus (Mouse) | PR |
| P70425 | Rit2 | GTP-binding protein Rit2 | Mus musculus (Mouse) | PR |
| Q8VEL9 | Rem2 | GTP-binding protein REM 2 | Mus musculus (Mouse) | PR |
| P62071 | Rras2 | Ras-related protein R-Ras2 | Mus musculus (Mouse) | PR |
| P36860 | Ralb | Ras-related protein Ral-B | Rattus norvegicus (Rat) | PR |
| P08644 | Kras | GTPase KRas | Rattus norvegicus (Rat) | SS |
| Q9WTY2 | Rem2 | GTP-binding protein REM 2 | Rattus norvegicus (Rat) | PR |
| P20171 | Hras | GTPase HRas | Rattus norvegicus (Rat) | SS |
| Q5BJQ5 | Rit2 | GTP-binding protein Rit2 | Rattus norvegicus (Rat) | PR |
| P97538 | Mras | Ras-related protein M-Ras | Rattus norvegicus (Rat) | PR |
| B7ZTR0 | cplane2 | Ciliogenesis and planar polarity effector 2 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| P79737 | nras | GTPase NRas | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| A1DZY4 | zgc:110179 | Ras-like protein family member 11A-like | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAAAGWRDGS | GQEKYRLVVV | GGGGVGKSAL | TIQFIQSYFV | TDYDPTIEDS | YTKQCVIDDR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AARLDILDTA | GQEEFGAMRE | QYMRTGEGFL | LVFSVTDRGS | FEEIYKFQRQ | ILRVKDRDEF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PMILIGNKAD | LDHQRQVTQE | EGQQLARQLK | VTYMEASAKI | RMNVDQAFHE | LVRVIRKFQE |
| 190 | 200 | ||||
| QECPPSPEPT | RKEKDKKGCH | CVIF |