P61981
Gene name |
YWHAG |
Protein name |
14-3-3 protein gamma |
Names |
Protein kinase C inhibitor protein 1, KCIP-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:7532 |
EC number |
|
Protein Class |
|
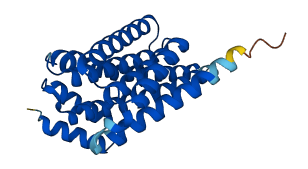
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

22 structures for P61981
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2B05 | X-ray | 255 A | A/B/C/D/E/F | 2-247 | PDB |
| 3UZD | X-ray | 186 A | A | 1-247 | PDB |
| 4E2E | X-ray | 225 A | A | 1-247 | PDB |
| 4J6S | X-ray | 308 A | A/B/C/D | 1-247 | PDB |
| 4O46 | X-ray | 290 A | A/B/C/D/E/F | 1-247 | PDB |
| 5D3E | X-ray | 275 A | A/B/E/F/I/J | 1-238 | PDB |
| 6A5S | X-ray | 210 A | A/B/D/G | 1-247 | PDB |
| 6BYJ | X-ray | 290 A | A/B/C/D/E/F | 2-241 | PDB |
| 6BYL | X-ray | 335 A | A/B/C/D/E/F | 2-241 | PDB |
| 6BZD | X-ray | 267 A | A/B/C/D | 2-247 | PDB |
| 6FEL | X-ray | 284 A | A/B/C/D | 1-234 | PDB |
| 6GKF | X-ray | 260 A | A/B/C/D/E/F/G/H | 1-234 | PDB |
| 6GKG | X-ray | 285 A | A/B/C/D/E/F/G/H | 1-234 | PDB |
| 6S9K | X-ray | 160 A | A | 1-234 | PDB |
| 6SAD | X-ray | 275 A | A/B | 1-234 | PDB |
| 6Y4K | X-ray | 300 A | A/B | 1-234 | PDB |
| 6Y6B | X-ray | 308 A | A/B | 1-234 | PDB |
| 6ZBT | X-ray | 180 A | A/B/C/D | 1-234 | PDB |
| 6ZC9 | X-ray | 190 A | A/B/C/D | 1-234 | PDB |
| 7A6R | X-ray | 270 A | A/B/C/D | 1-234 | PDB |
| 7A6Y | X-ray | 250 A | A/B/C/D | 1-234 | PDB |
| AF-P61981-F1 | Predicted | AlphaFoldDB |
109 variants for P61981
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
VAR_080224 CA367758305 RCV000505697 rs1554618767 |
15 | E>A | Developmental and epileptic encephalopathy, 56 DEE56; unknown pathological significance [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
VAR_080225 CA367778044 rs1554616652 RCV000664053 |
50 | K>Q | Developmental and epileptic encephalopathy, 56 found in an individual with autism; unknown pathological significance [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000816037 rs1583981736 RCV002267626 CA367778002 COSM73324 RCV001788360 |
57 | R>C | Developmental and epileptic encephalopathy, 56 ovary Microcephaly [ClinVar, Cosmic] | Yes |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
|
RCV000505692 rs1554616630 VAR_080226 RCV002524425 CA367777399 |
129 | D>E | Developmental and epileptic encephalopathy, 56 DEE56 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA367777361 rs1554616628 RCV000505695 RCV001591150 VAR_080227 |
132 | R>C | Developmental and epileptic encephalopathy, 56 DEE56 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1583981615 RCV001171618 CA367777355 RCV000995916 |
132 | R>H | Developmental and epileptic encephalopathy, 56 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1554616627 VAR_080228 CA367777347 RCV000664047 |
133 | Y>S | Developmental and epileptic encephalopathy, 56 probable disease-associated variant found in an individual with neurodevelopmental disorder [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA367777137 rs1389455796 RCV000987899 |
151 | E>* | Developmental and epileptic encephalopathy, 56 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
RCV001266968 rs1803516480 |
184 | Y>* | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1803513587 RCV001328674 |
228 | D>V | Developmental and epileptic encephalopathy, 56 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1176167493 CA367758445 |
6 | Q>E | No |
ClinGen Ensembl |
|
|
rs1235900410 CA367758440 |
6 | Q>R | No |
ClinGen gnomAD |
|
|
CA4306610 rs780642467 |
11 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA4306611 rs566717170 |
11 | A>T | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 14 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA367758313 rs1320970802 |
14 | A>V | No |
ClinGen TOPMed |
|
|
CA4306608 rs746573987 |
19 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA367758224 rs1290226481 |
20 | Y>C | No |
ClinGen gnomAD |
|
|
rs371405636 CA4306606 |
21 | D>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA367758210 rs1344323633 |
22 | D>N | No |
ClinGen gnomAD |
|
|
CA4306604 rs764951861 |
23 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1426519806 CA367758203 |
23 | M>V | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1275951666 CA367758186 |
25 | A>G | No |
ClinGen gnomAD |
|
|
CA367758183 rs1411908821 |
26 | A>T | No |
ClinGen gnomAD |
|
|
rs759666738 CA367758155 CA4306600 |
29 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs1430500844 CA367778162 |
32 | E>Q | No |
ClinGen gnomAD |
|
|
CA4306568 rs771049818 |
36 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA367778025 rs1313967468 |
52 | V>A | No |
ClinGen gnomAD |
|
|
CA4306563 rs780366000 |
52 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs758388896 CA4306562 |
53 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA160927684 rs866370817 |
55 | A>T | No |
ClinGen Ensembl |
|
|
CA4306561 rs750576954 |
56 | R>C | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 65 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs769855430 CA160927673 |
66 | I>T | No |
ClinGen Ensembl |
|
|
CA367777904 rs1403141077 |
70 | T>R | No |
ClinGen gnomAD |
|
|
CA160927650 rs1057100383 |
74 | G>S | Variant assessed as Somatic; 4.624e-05 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1161419188 CA367777840 |
79 | I>M | No |
ClinGen TOPMed |
|
|
CA367777830 rs1439613878 |
81 | M>V | No |
ClinGen gnomAD |
|
|
CA4306556 rs760331444 |
83 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA160927634 rs1037532353 |
84 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs758954176 CA4306553 |
86 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA367777797 rs1563661628 |
86 | R>W | No |
ClinGen Ensembl |
|
|
rs770925257 CA4306551 |
89 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA367777721 rs1272649053 |
97 | C>R | No |
ClinGen TOPMed |
|
|
rs1400963948 CA367777710 |
98 | Q>P | No |
ClinGen gnomAD |
|
|
CA4306550 rs759611694 COSM1755360 RCV000902447 |
99 | D>N | urinary_tract [Cosmic] | No |
ClinGen cosmic curated ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA367777684 rs1307223884 |
102 | S>N | No |
ClinGen gnomAD |
|
|
rs762781768 CA160927604 |
106 | N>D | No |
ClinGen Ensembl |
|
|
CA160927595 rs984280298 |
106 | N>K | No |
ClinGen Ensembl |
|
|
rs773103074 CA4306549 |
106 | N>S | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 110 | K>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1394668720 CA367777623 |
111 | N>I | No |
ClinGen gnomAD |
|
|
CA367777616 rs1387672498 |
112 | C>Y | No |
ClinGen gnomAD |
|
|
rs1419668779 CA367777604 |
114 | E>Q | No |
ClinGen gnomAD |
|
| TCGA novel | 116 | Q>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 119 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs745872119 CA367777516 |
120 | K>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs745872119 CA4306544 |
120 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs921555508 CA367777468 |
123 | Y>* | No |
ClinGen TOPMed |
|
|
rs976962773 CA160927549 |
126 | M>L | No |
ClinGen Ensembl |
|
|
rs754370006 CA4306541 |
138 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1246308613 CA367777277 |
139 | T>S | No |
ClinGen TOPMed |
|
|
CA160927515 rs989458518 |
141 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1224824021 CA367777240 |
142 | K>R | No |
ClinGen gnomAD |
|
|
CA4306538 rs753162401 |
143 | R>K | No |
ClinGen ExAC gnomAD |
|
|
CA367777222 rs1283661746 |
144 | A>T | No |
ClinGen gnomAD |
|
|
rs776692453 CA4306537 |
144 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1270651 CA4306535 rs773949985 |
145 | T>M | oesophagus [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA367777192 rs1385789611 |
146 | V>G | No |
ClinGen TOPMed |
|
|
rs1389455796 CA367777136 |
151 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs747894498 CA4306530 |
155 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA367777068 rs111385409 |
155 | S>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA367777062 rs1333026759 |
156 | E>K | No |
ClinGen TOPMed |
|
|
rs746119956 CA4306527 |
159 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA367777002 rs746746074 |
160 | I>N | No |
ClinGen TOPMed |
|
|
CA160927471 rs746746074 |
160 | I>T | No |
ClinGen TOPMed |
|
| TCGA novel | 161 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4306526 rs547605838 |
162 | K>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
| TCGA novel | 163 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1270980685 CA367776937 |
165 | M>V | No |
ClinGen gnomAD |
|
|
CA4306525 rs757293780 |
166 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA367776893 rs1583981528 |
168 | T>P | No |
ClinGen Ensembl |
|
|
CA367776873 rs1583981523 |
169 | H>P | No |
ClinGen Ensembl |
|
|
CA367776852 rs1289299592 |
171 | I>V | No |
ClinGen gnomAD |
|
|
CA367776831 rs1208117178 |
172 | R>L | No |
ClinGen gnomAD |
|
| TCGA novel | 174 | G>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1422674540 CA367776789 |
176 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1218412501 CA367776781 |
176 | A>V | No |
ClinGen gnomAD |
|
| TCGA novel | 182 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1330999969 CA367776678 |
184 | Y>C | No |
ClinGen gnomAD |
|
|
rs867451102 CA160927440 |
186 | I>T | No |
ClinGen Ensembl |
|
|
CA4306518 rs532533392 |
187 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4306516 rs762541714 |
189 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA4306514 rs765021527 |
190 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA160927414 rs897103736 |
194 | C>R | No |
ClinGen TOPMed |
|
| TCGA novel | 197 | A>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1563661472 CA367776477 |
199 | T>A | No |
ClinGen Ensembl |
|
|
rs749364591 CA4306507 |
200 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA4306502 rs755349826 |
206 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs370378730 CA4306499 |
209 | D>E | No |
ClinGen ESP ExAC gnomAD |
|
|
rs749898700 CA4306498 |
211 | L>F | No |
ClinGen ExAC |
|
|
CA367776058 rs1303116971 COSM1240675 |
231 | T>M | oesophagus [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1178065393 CA367776042 |
234 | T>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs1333221474 CA367776037 |
234 | T>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA367776032 rs1236846905 |
235 | S>T | No |
ClinGen TOPMed |
|
|
rs769355192 CA4306485 |
241 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1425390006 CA367775972 |
243 | G>S | No |
ClinGen gnomAD |
|
|
CA367775962 rs1307963964 |
244 | E>K | No |
ClinGen gnomAD |
|
| TCGA novel | 248 | N>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
1 associated diseases with P61981
[MIM: 617665]: Developmental and epileptic encephalopathy 56 (DEE56)
A form of epileptic encephalopathy, a heterogeneous group of severe early-onset epilepsies characterized by refractory seizures, neurodevelopmental impairment, and poor prognosis. Development is normal prior to seizure onset, after which cognitive and motor delays become apparent. DEE56 is an autosomal dominant condition. {ECO:0000269|PubMed:28777935}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A form of epileptic encephalopathy, a heterogeneous group of severe early-onset epilepsies characterized by refractory seizures, neurodevelopmental impairment, and poor prognosis. Development is normal prior to seizure onset, after which cognitive and motor delays become apparent. DEE56 is an autosomal dominant condition. {ECO:0000269|PubMed:28777935}. Note=The disease is caused by variants affecting the gene represented in this entry.
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| focal adhesion | A cell-substrate junction that anchors the cell to the extracellular matrix and that forms a point of termination of actin filaments. In insects focal adhesion has also been referred to as hemi-adherens junction (HAJ). |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| identical protein binding | Binding to an identical protein or proteins. |
| insulin-like growth factor receptor binding | Binding to an insulin-like growth factor receptor. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase C binding | Binding to protein kinase C. |
| protein kinase C inhibitor activity | Binds to and stops, prevents or reduces the activity of protein kinase C, an enzyme which phosphorylates a protein. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| cellular response to insulin stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an insulin stimulus. Insulin is a polypeptide hormone produced by the islets of Langerhans of the pancreas in mammals, and by the homologous organs of other organisms. |
| negative regulation of protein kinase activity | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase activity. |
| protein localization | Any process in which a protein is transported to, or maintained in, a specific location. |
| protein targeting | The process of targeting specific proteins to particular regions of the cell, typically membrane-bounded subcellular organelles. Usually requires an organelle specific protein sequence motif. |
| regulation of neuron differentiation | Any process that modulates the frequency, rate or extent of neuron differentiation. |
| regulation of signal transduction | Any process that modulates the frequency, rate or extent of signal transduction. |
| regulation of synaptic plasticity | A process that modulates synaptic plasticity, the ability of synapses to change as circumstances require. They may alter function, such as increasing or decreasing their sensitivity, or they may increase or decrease in actual numbers. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
37 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P27348 | YWHAQ | 14-3-3 protein theta | Homo sapiens (Human) | PR |
| Q04917 | YWHAH | 14-3-3 protein eta | Homo sapiens (Human) | PR |
| P63104 | YWHAZ | 14-3-3 protein zeta/delta | Homo sapiens (Human) | PR |
| P31946 | YWHAB | 14-3-3 protein beta/alpha | Homo sapiens (Human) | PR |
| P49106 | GRF1 | 14-3-3-like protein GF14-6 | Zea mays (Maize) | PR |
| P68510 | Ywhah | 14-3-3 protein eta | Mus musculus (Mouse) | PR |
| P63101 | Ywhaz | 14-3-3 protein zeta/delta | Mus musculus (Mouse) | PR |
| P93784 | HOX | 14-3-3-like protein 16R | Solanum tuberosum (Potato) | PR |
| Q41418 | 14-3-3-like protein | Solanum tuberosum (Potato) | PR | |
| P63102 | Ywhaz | 14-3-3 protein zeta/delta | Rattus norvegicus (Rat) | PR |
| Q6EUP4 | GF14E | 14-3-3-like protein GF14-E | Oryza sativa subsp japonica (Rice) | PR |
| Q06967 | GF14F | 14-3-3-like protein GF14-F | Oryza sativa subsp japonica (Rice) | PR |
| Q7XTE8 | GF14B | 14-3-3-like protein GF14-B | Oryza sativa subsp japonica (Rice) | PR |
| Q6ZKC0 | GF14C | 14-3-3-like protein GF14-C | Oryza sativa subsp japonica (Rice) | PR |
| Q84J55 | GF14A | 14-3-3-like protein GF14-A | Oryza sativa subsp japonica (Rice) | PR |
| Q2R2W2 | GF14D | 14-3-3-like protein GF14-D | Oryza sativa subsp japonica (Rice) | PR |
| Q2R1D5 | GF14H | Putative 14-3-3-like protein GF14-H | Oryza sativa subsp japonica (Rice) | PR |
| Q96450 | GF14A | 14-3-3-like protein A | Glycine max (Soybean) (Glycine hispida) | PR |
| Q96453 | GF14D | 14-3-3-like protein D | Glycine max (Soybean) (Glycine hispida) | PR |
| Q96452 | GF14C | 14-3-3-like protein C | Glycine max (Soybean) (Glycine hispida) | PR |
| Q9C5W6 | GRF12 | 14-3-3-like protein GF14 iota | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9S9Z8 | GRF11 | 14-3-3-like protein GF14 omicron | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P46077 | GRF4 | 14-3-3-like protein GF14 phi | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q01525 | GRF2 | 14-3-3-like protein GF14 omega | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q96300 | GRF7 | 14-3-3-like protein GF14 nu | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42645 | GRF5 | 14-3-3-like protein GF14 upsilon | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42644 | GRF3 | 14-3-3-like protein GF14 psi | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q96299 | GRF9 | 14-3-3-like protein GF14 mu | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P42652 | TFT4 | 14-3-3 protein 4 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93212 | TFT7 | 14-3-3 protein 7 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93209 | TFT3 | 14-3-3 protein 3 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93207 | TFT10 | 14-3-3 protein 10 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93211 | TFT6 | 14-3-3 protein 6 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93206 | TFT1 | 14-3-3 protein 1 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93208 | TFT2 | 14-3-3 protein 2 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93213 | TFT8 | 14-3-3 protein 8 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| P93214 | TFT9 | 14-3-3 protein 9 | Solanum lycopersicum (Tomato) (Lycopersicon esculentum) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MVDREQLVQK | ARLAEQAERY | DDMAAAMKNV | TELNEPLSNE | ERNLLSVAYK | NVVGARRSSW |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RVISSIEQKT | SADGNEKKIE | MVRAYREKIE | KELEAVCQDV | LSLLDNYLIK | NCSETQYESK |
| 130 | 140 | 150 | 160 | 170 | 180 |
| VFYLKMKGDY | YRYLAEVATG | EKRATVVESS | EKAYSEAHEI | SKEHMQPTHP | IRLGLALNYS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VFYYEIQNAP | EQACHLAKTA | FDDAIAELDT | LNEDSYKDST | LIMQLLRDNL | TLWTSDQQDD |
| DGGEGNN |